There are two predominant views regarding the genesis and development of humankind which differ markedly. What if a bridge could be built which joined them together not as two opposing views running distantly parallel, but rather along the exact same line? Evolution confidently states there is no Creator; with humans the random result of millions of years of rather coincidental mutations occurring with seemingly impossible odds. Scientists like atheists, eliminate a spiritual component from the equation of biogenesis, relying on a purely physical explanation. Creationists in the main, teach that a divine God created everything by His Spirit; yet accounting for an impossible human history condensed within a chronology of a mere six millennia.
What if the two could be married together? What if the time frames involved for mankind’s evolution is not millions or only thousands of years, but instead tens of thousands of years? With the Solar System and the Universe beyond, hundreds of thousands of years old? What if the beginning of life is explained through supernatural means, yet the physical dimensions constructing our world are a creation within a creation? What if a supreme Creator is the first source, but other beings have been responsible for preternatural interference and genetic manipulation in the incredible project here on planet Earth? It would mean that evolution is a viable theory, yet just not in the way scientists might think – refer article: Chance Chaos or Designated Design? Likewise, creation is a credible answer, though not quite in the manner Christians may imagine – refer Chapter XXII Alpha & Omega.
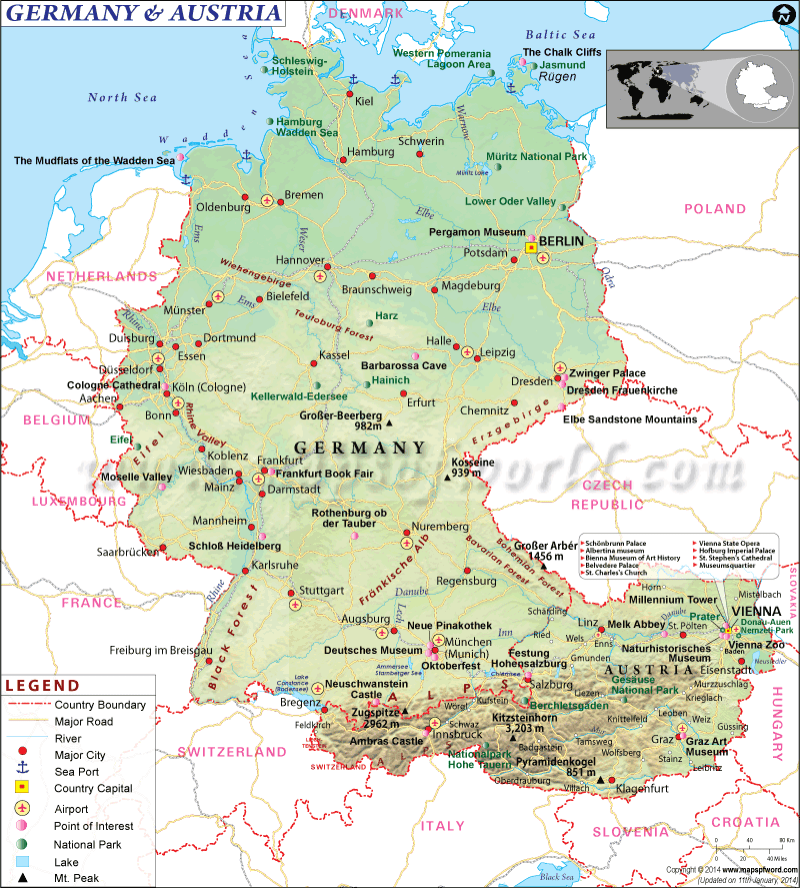
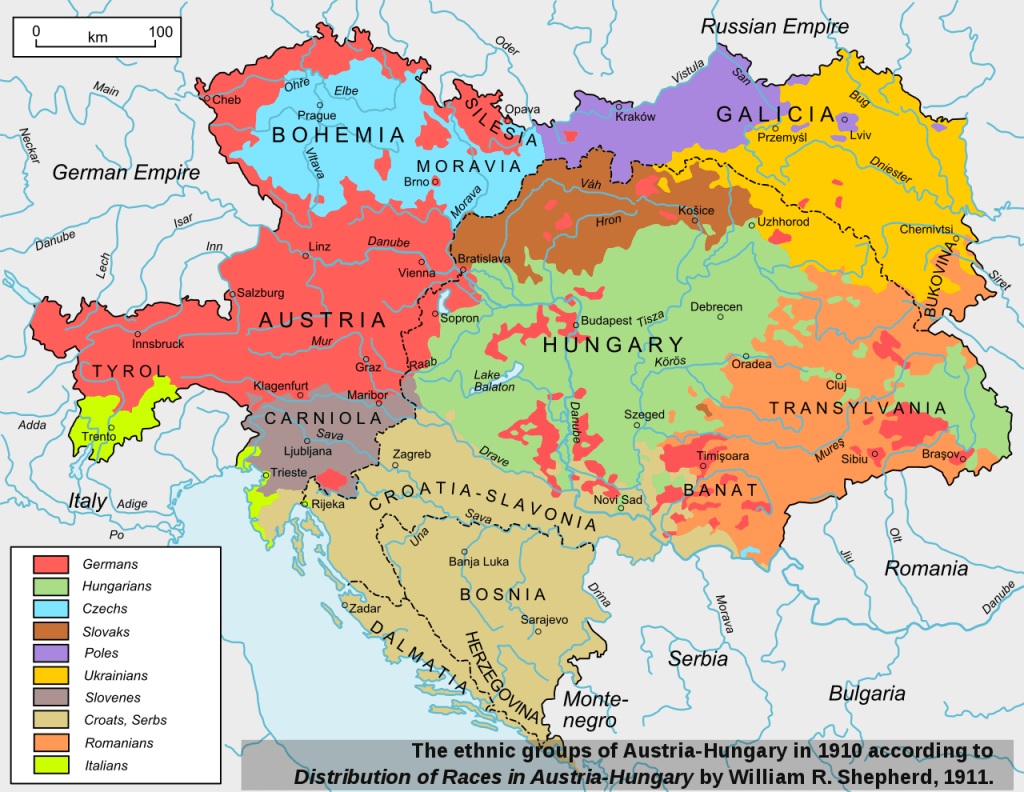
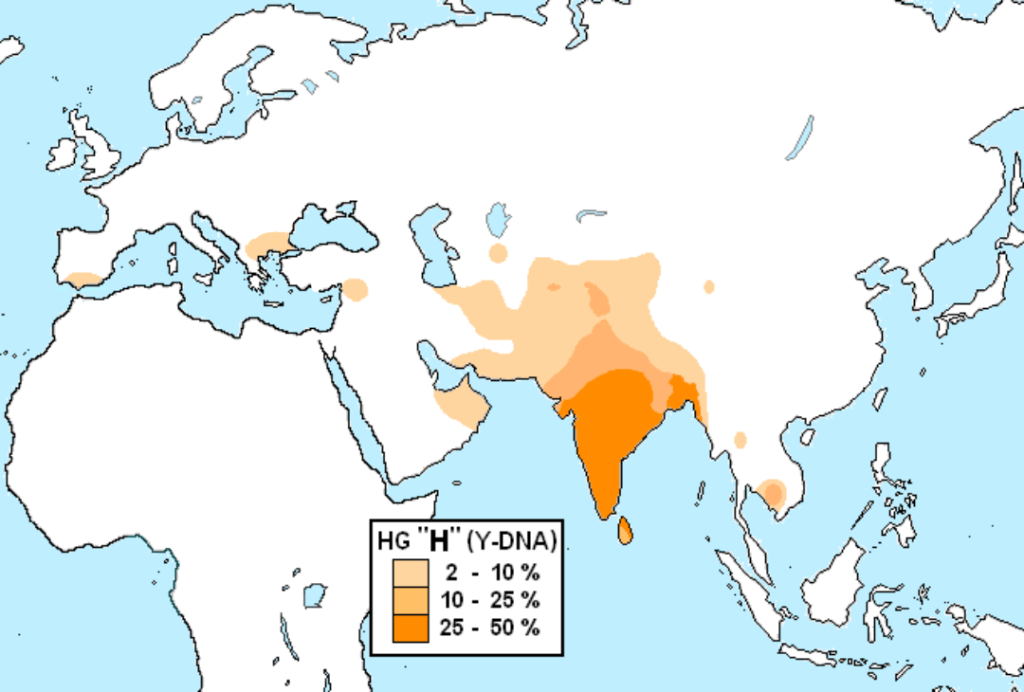
We will focus on the scientific field of genetics, principally Haplogroups and the evidence they provide for ancient lineal lines of descent through to our present day, resulting in the vast family tree that is the world’s two hundred countries, containing thousands of ethnicities. In turn, we will consider humanities ancestors who are named in the Bible and any credence to the three (actually four) broad ancestry groups and the sixteen (in reality twenty-one) major races descending from them. For those readers who may take umbrage with the word race, please refer to the concluding Chapter finalis verbum.
A Haplogroup is a series of mutations found in a chromosome. Specifically, they are a combination of alleles or Haplotypes and though located at different chromosomal regions, they are closely linked and tend to be inherited together. An allele (or allelomorph) may occur in pairs, or in multiple alleles which influences the expression or phenotype of any given human trait. The combination of alleles that in this instance a human carries, constitutes their genotype – the chemical composition of their personal DNA. Therefore, Haplogroups are detectable in the DNA of an individual and reveal with who, they share a common paternal or maternal ancestor.
Haplogroups are normally identified by a code comprising an initial letter of the alphabet, and ‘refinements consist of additional number and letter combinations, such as’ for example R → R1 → R1a and R1b, or R2 and so forth. This simplifies genealogical tracing of the genetic mutations.
In human genetics, the Haplogroups studied are: the patrilineal line consisting of a Y-DNA Haplogroup from a Y sex chromosome passed only from fathers to sons and the matrilineal line comprising a mtDNA Haplogroup consisting of mitochondria passed from mothers to offspring of both sexes. Females inherit an X chromosome from both their mother and father and have two X chromosomes. Therefore, females are XX. Conversely, males inherit one X chromosome from their mothers and a Y chromosome from their fathers. Thus, males are XY.
Haplogroups define every ethnicity or racial strand of descent. Haplogroups are split between the genetic information received from ones mother, mtDNA and from ones father, Y-DNA. The proviso is that a daughter does not receive the Y chromosome Haplogroup from her father. Whereas a son receives both the mitochondrial DNA from his mother and the Y chromosome from his father. Thus, there are two sets of Haplogroups for males.
Y-DNA is how paternal Haplotypes are inherited through a direct ancestral male line for countless generations. Because females do not inherit Y-DNA, they do not possess a paternal Haplogroup. Whereas maternal Haplogroup information, including for males is found within the mitochondria of our cells; hence the term mitochondrial DNA or mtDNA. ‘Mitochondria are small organelles that lie in the cytoplasm of eukaryotic cells… Their primary function is to provide energy to the cell.’ Scientists are not exactly sure why mitochondrial DNA is not passed down from fathers. Neither recombines and both Y-DNA and mtDNA change only by chance mutation at each generation with no intermixture between parents’ genetic material.
The remainder of about 98% of an individual’s genetic material – other than the two sex chromosomes X and Y, inherited from both parents – are autosomal chromosomes. These contain segments of DNA that a person shares with everyone they are related to. In essence, the fundamental difference between autosomal DNA and Haplogroup DNA is that the latter provides a genetic snapshot. One that follows a single line of your father’s fathers and mother’s mothers, revealing Haplogroup sub-clades which formed hundreds or thousands of years ago.
Though Haplogroups only provide surface ancestral information for an individual and are not as comprehensive as autosomal results; they do reveal ancient origins of ethnicities and shared common ancestry. Autosomal DNA concentrates on traces from perhaps five to ten generations back in time, over a few centuries but in so doing includes all of ones ancestral branches in its scope, including not just their father’s father, but ones mother, her mother and father and so forth; providing more detail about a person’s personal and immediate ancestry.
An enumeration of the chemical process in analysing Y-DNA and mtDNA Haplogroups is provided in the following article:
The Genetic Origin of the Nations, Christian Churches of God, 2006 & 2020 – emphasis mine:
‘The YDNA and mtDNA are measured in two different ways. YDNA is measured in what are termed polymorphs. These polymorphs are allocated a numeric value and, according to the value when tested, the sub-groupings that are formed are called clades and subclades of the overall grouping which is called a Haplogroup. These values record the change in YDNA mutations and lines. The YDNA system that has been allocated to the male human species is grouped into a series of Haplogroups from A to R. The usual extensive measurement (using the Arizona system) is usually of 37 sites as markers. Basic testing is done for the first twelve, then to 25, and then on to 37 of these polymorphic sites, or locations to determine relatedness and Haplogroup association. There could be some 100 or more markers tested for changes (a.k.a. polymorphisms).’

‘The mitochondria, first sequenced in 1981, became known as the Cambridge Reference Sequence (CRS). The CRS has been used as a basis for comparison with individual mtDNA. In other words, any place in an individual mtDNA that has a difference from the CRS is characterized as a mutation. If a result shows no mutations at all it means that the mtDNA matches the CRS. A mutation happens when: a) a base replaces another base – for example a C (Cytosine) replaces an A (Adenine); b) a base is no longer in that position, or a deletion; and c) a new base is inserted between the other bases without replacing any other (an insertion).
The mtDNA is determined by reporting the polymorphic site such as for example 311C, meaning a mutation has occurred at base pair 16,311 and the base that changed here was actually changed to cytosine. The number 16,000 is the commencement point for DNA numbering and thus the 16,000 is dropped and the numbers used are the numbers in excess. So 16,311 becomes 311 and the letter indicates the chemical at that point in the polymorph. It is this change of the polymorphic site that determines the genetic ancestry, as the parent passes on to the offspring the DNA polymorphisms that they have with the same or similar numerical values. When tested, these values that are not exactly the same as the parent are termed mutations. The values thus vary and have determined the tribal groupings of the world’s nations.’
The scientific confirmation of an original female Homo sapiens progenitor is discussed in the following quote. Mitochondrial Eve was the name chosen by researchers for the woman who is understood to be the most recent common female lineal ancestor of all living humans.
Gods of the New Millennium, Alan Alford, 1997 – emphasis & bold mine:
‘In 1987, Allan Wilson, Mark Stoneking and Rebecca Cann, from the University of California at Berkeley, declared that all women alive today must have had a common genetic ancestor… How did they arrive at this conclusion? This… has been made possible by the discovery of mitochondria the tiny bodies within a cell that are responsible for production of energy through breakdown of sugars. Unlike our other DNA, which is scrambled by sexual recombination, mitochondrial DNA (mtDNA) is inherited virtually unchanged through the female line and is thus a perfect marker to trace ancestral relations. Moreover, it mutates at a predictable rate. The number of differences between the mtDNA in a worldwide sample of 135 different women allowed Wilson, Stoneking and Cann to compare how far back the ancestors of these women had diverged.
In order to calibrate the divergences, the researchers used a comparison of mtDNA between man and chimpanzees, based on a separation 5 million years ago. And that led to the conclusion that a common ancestor named “Mitochondrial Eve” must have lived 250-150,000 years ago. This genetic evidence has been challenged, due to its calibration with the chimpanzees, whose separation date from man is not known with certainty. As Richard Dawkins has pointed out, this does not mean that Eve was the only woman on Earth at that time, just that she is the only one who has an unbroken line of female descendants. The chances are that many earlier Eves have descendants alive today, but their ancestry has passed, at some point, through the male line only. Despite the new mtDNA dates, most studies still tend to support and cite the 200,000 BP common ancestor.’
Constant readers and those who have read Appendix IV: An Unconventional Chronology will be aware the time frame for Adam and Eve may be as recent as thirty-thousand years ago. The supposed separation between chimps and man is not a given let alone the dating proposed. The purpose is not to discuss or digress on topics discussed in preceding chapters and articles. These include Homo erectus, Neanderthal man, the creation of Homo sapiens and the origin of Adam and Eve. Please refer to Chapter I Noah Antecessor Nulla; Chapter XVI Shem Occidentalis; Chapter XXII Alpha & Omega; and articles: Homo neanderthalensis I, II, III & IV.

The above trees for patrilineal Y-DNA and matrilineal mtDNA are reasonably accurate.
First, an introduction to each of the Haplogroups and a synopsis for each, while presenting where applicable Noah’s sons and grandsons; matching where we can the Y-DNA Haplogroups and mtDNA with Noah’s sons wives.
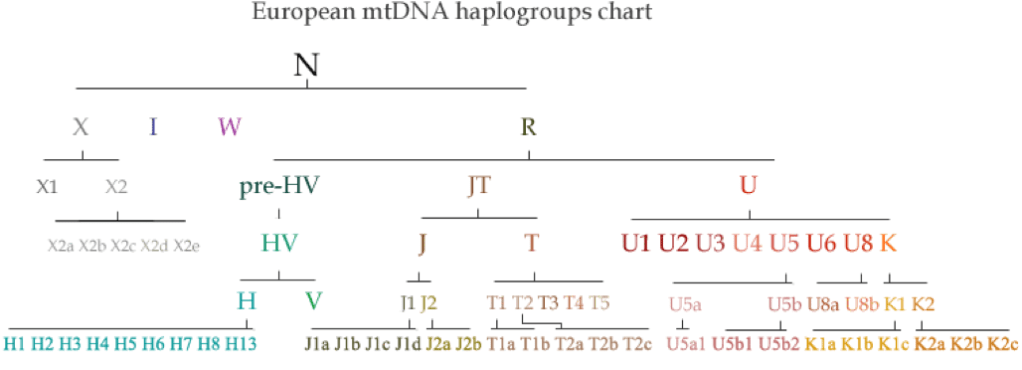
Maternal mtDNA Haplogroups are lettered as such:
A, B, C, D, E, F, G, H, HV, I, J, K, L0, L1, L2, L3, L4, L5, L6, M, N, P, Q, R, S, T, U, V, W, X, Y, and Z.
Haplogroups are used to define genetic populations invariably explained from either a chronological origin or a geographic orientation. The following are recognised divisions for mtDNA Haplogroups:
African:
L0, L1, L2, L3, L4, L5, L6
West Eurasian – including North Africa, the Middle East and South Asia:
H, T, U, V, X, K, I, J, N, R, W
East Eurasian:
A, B, C, D, E, F, G, M, Y, Z
Native American:
A, B, C, D, X
Austronesian-Melanesian:
P, Q, S
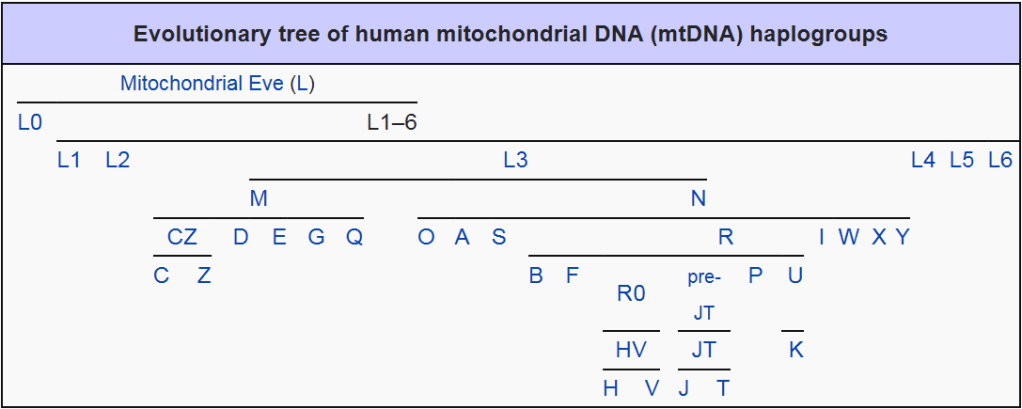
Mitochondrial Haplogroups are divided into three main groups, designated by the sequential letters L, M and N. Early Homo sapiens first split within the L group between L0 and L1 to L6. Haplogroups L1 to L6 ‘gave rise to other L groups, one of which, L3, split into the M and N [groups].’ The M group comprises the first lineages of Haplogroup M found throughout Asia, the Americas, Melanesia, parts of the Horn of Africa and North Africa.
The N Haplogroup is thought to represent another later macro-lineage. This maternal line split into another group called R. ‘Haplogroup R consists of two subgroups defined on the basis of their geographical distributions, one found in southeastern Asia and Oceania and the other containing almost all of the modern European populations. Haplogroup N(xR), [that is] mtDNA that belongs to the N group but not to its R subgroup, is typical of Australian aboriginal populations, while also being present at low frequencies among many populations of Eurasia and the Americas.’
Haplogroup L comprises nearly all sub-Saharan Africans. L0 is the most ancient mtDNA Haplogroup. L1 is the next oldest branch of the maternal family tree, being a daughter of the mitochondrial Eve Haplogroup L and a sister to L0. It is most frequently found in western and central sub-Saharan Africa; seldom appearing in eastern or southern Africa. It is L1 which is the ancestor to branches L2 to L6.
Fascinatingly, it was the group L3 mutations which gave rise to all the non-African Haplogroups found today in both West Eurasians and East Eurasians. Haplogroup L3 comprises some 40% of the sub-Saharan maternal variation. L2 is found in a third of sub-Saharan Africans. Its subgroup L2a, is not only the most common mtDNA Haplogroup among African Americans, but is the most frequent and widespread mtDNA cluster in Africa. For further in-depth discussion on the L0-L6 Haplogroups, refer Chapter XII Canaan & Africa.
The Genetic Origin of the Nations, 2006 & 2020:
“Noah was understood to be pure in his generations. The Bible also maintains that the people in the Ark were all the family of Noah. Thus, to properly account for the genetic diversity, Noah must have maintained the capacity to throw genetically distinct offspring, and this offspring had the characteristics of the line from which it came, but not the entire sequence that Noah had originally. For Noah to be the father of the human structure he is held to have had the capacity for the… YDNA substructure, as all humans are descended from him. Any male on the planet will have only the mutations that signify his branch and path. Noah held the base YDNA that was able to mutate into… other subgroups.
… when we examine the tree of mtDNA we find some interesting group derivatives. The so-called “supergroups” are really only in three basic groups. In other words, they came from three main female lines. That is what we would expect to find if we assume there were only three females that bred on from the Ark, namely the wives of Shem, Ham and Japheth. These Haplogroups are all descended from a single female supergroup, namely Haplogroup L. So in reality, all females are descended from one female line, Hg L. That is super L. This line then split into L1, and then L2 and L3. The line L3 diverged and from L3 came the other mtDNA mutations. Thus, all females came from one Eve whose mtDNA line was L.
The supergroups M and N were next to diverge or mutate. From a biblical point of view we can argue easily that L was formed with Eve and the other groups were pre-Flood divisions that came on to the Ark. Thus, we could correctly argue that L, M, and N came on to the Ark within the accepted biblical account. All mtDNA Haplogroups are subdivisions of L, then M and N and subsequently R, which itself is a mutation of Hg N.
Thus, we can assume that Eve produced the line L and the three wives of Shem, Ham and Japheth are at least the three groups L, M and N. There may have been further divisions given the fact that Noah may have had daughters not mentioned and their mtDNA line may have been L, or M or N. It may have even been R, if we assume that the entire L line came in through the wife of Ham, as the L line is almost confined to the sub-Saharan tribes.
We also have to address the fact that Eve was dark skinned… Thus the capacity for the development of skin colour was an original trait [even if recessive] of the human creation.
M produced three subdivisions… including C [and Z, which split from each other], and D and G… [with subdivisions] E and Q… [all associated with East Asian peoples].
We might thus also deduce that the wives of the sons of Noah were taken from the one family lineage, maintaining purity in the generations in the female line also. The L2 and L3 split may have come from the family structure before the Flood. [Any] daughters of Noah and the wives of the sons could have carried all three of the L subdivisions and the basic core sub-groups of M, N and perhaps R. It is therefore possible that the women of the Ark… could easily have contained the basis for the modern mtDNA diversity.
The supergroup N… split… [including] Haplogroups I and W… The R supergroup split into the following: B; F; HV, which split into H and V; P; The J and T subdivision; and U, from which came K… [all associated with European peoples].”
According to the author, the mtDNA super Haplogroup L originated with Eve… and split into (L0) L1, L2 and L3. All mtDNA L Haplogroups from L0 to L6 are primarily associated with Black African people and to a lesser extent, Arabs. The remainder of the mtDNA Haplogroups then derived or mutated from L3. L3 gave rise to the super subgroups M and N. Broadly speaking, L3 relating to African peoples; M with East Asian; and N with Europeans. The author states that Japheth, Ham and Shem’s wives would have carried these new mutations. For the three wives of Noah’s sons to each represent these three core racial strands, the connecting dots not suggested by the author are that these wives could have also been daughters of Noah by his wife Emzara. Though there is reason to believe this is not the case.
Noah would have passed on to each son the paternal genetic sequencing (Y chromosome DNA) for Japheth and his subsequent seven sons; Ham and his three sons; Shem and his five sons; and finally Canaan and his six sons – refer Chapter XI Ham Aequator; and Chapter XII Canaan & Africa. Noah’s wife would have received the maternal recessive genes (Mitochondria DNA) originating in the L3 line from Eve, which included Haplogroups M and N. Thus, L3, M and N were new mutations that had not existed during the antediluvian epoch. The new Haplogroups had lain dormant until being activated or awakened by congress with Noah.
The new racial characteristics could have been carried by Noah’s daughters (in law), ‘Adataneses, Na’eltama’uk and Sedeqetelebab who then married respectively, Japheth, Ham and Shem whose descendants would exhibit the new mutations, revealing two new racial strands – bluntly and broadly: yellow from Japheth (C, D) and ‘Adataneses (M); and white from Shem (G) and Sedeqetelebab (N, R); to add to an original brown skin tone. The latter now carried a new mutation too; creating extra diversity in Ham (H, J) and Na’eltama’uk (L0, L1-L6). Canaan (A, B, E) is a separate line again and is discussed in depth in Chapter XI Ham Aequator; and Chapter XII Canaan & Africa.
What is of fascinating interest is that while the white line when it mutated long after the Flood was new; the yellow line of descent was a throwback to the people of Day Six – refer articles: Homo neanderthalensis I, II, III & IV; and Chapter II Japheth Orientalium.

‘Adataneses
It is understandable why these eight people were saved and that not just Noah was genealogically pure, but so was his wife. They then had (probably) three to (possibly) six children prior to the flood who received the genetic sequencing for the three (actually four) new core racial lines, which then mutated into the sixteen (in reality twenty-one) new sub-racial strands through their children after the flood – Noah’s and Emzara’s grandchildren.
This leaves the L and specifically the L0 pre-flood line from Eve. The simple answer is that L was passed to Cain and his family line and L0 was passed to Seth and his line of descent which later included Noah and his wife. L3 with M and N, being the later mutations from Seth’s line L0 after the flood. The L and L0 lines were mid-toned skinned lines, with the darker and lighter shades of skin and racial diversity included in the L3 line we presently have now, deriving from Noah’s descendants. The undeniable scientific support for this argument, is that a black couple can have white children, but a white couple cannot have a black child.

Science confirms white skin is a mutation – the SLC24A5 gene on Chromosome 15 – of an originally darker human. For instance, East Asians have acquired mutations in other genes which result in lighter skin, while retaining black hair. The gene mutation SLC24A5 changes just one building block in the protein, contributing about a third of the pigment loss that makes black skin white; accounting for the differences in skin tone between peoples of African and European ancestry for example – refer Chapter XVI Shem Occidenatlis.

Sedeqetelebab
The fact of the matter is that everyone descends from mutated DNA genetic code which originally began with ancestors of the distant past. So which peoples today are they most closely aligned with? The oldest Haplogroup from the mtDNA tree passed from mothers to sons and daughters, originating from mitochondrial Eve, is L0. This Haplogroup is indicative of the peoples of Southern Africa. The Khoisan are a good example and they possess a light brown skin. Thus ‘Eve’ would have been in all probability… light brown – refer Chapter XII Canaan & Africa.
It is important to understand that recessive genetic information is shared amongst family members, sometimes in surprising ways. Just as cousins can be more alike and develop a closer bond than with their own siblings. What is significant is that the mtDNA Haplogroup L, though shared in common origin by Europeans and East Asians through Shem, Japheth and their wives ‘Adataneses and Sedeqetelebab in the form of the L3 mutation, it was the L0 to L6 mutations which have been overwhelmingly carried by Noah’s^ fourth and youngest son, Canaan.
Coupled with this, is that as the macro-Haplogroup M is almost exclusively associated with East Asians, macro-Haplogroup N is not just the domain of Europeans descended from Shem but also for Arabs, Indians and Pakistanis descended from Ham. The most logical answer for this occurrence is that Ham’s sons – Cush, Mizra and Phut – must have taken wives from Shem’s line and hence why they share mtDNA in common with them.
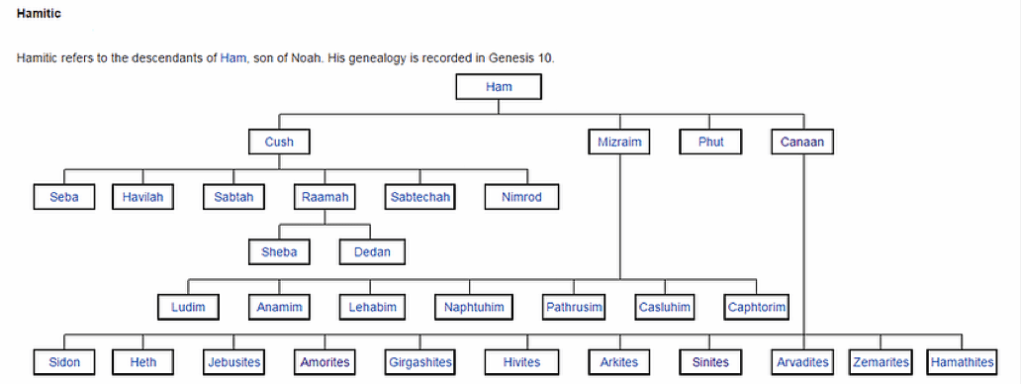
Following L, the next major maternal Haplogroup branch is M. M1 intrigues scientists with its presence in Ethiopian, Somali and Indian populations, where M3 is located. These are descendants of Canaan’s sons and from Ham’s son Cush. What may have a bearing is that Ham’s wife Na’eltama’uk is the mother of both Canaan and Cush who share different fathers.^

Na’eltama’uk
Yet the Haplogroup mutations stemming from M as follows, are all defining maternal markers for sons from Japheth. Haplogroups derived from super group M include: CZ found in Siberians; with branch C found amongst Amerindians; while branch Z is carried by the Saami; and minimally in Korean, North Chinese and Central Asian populations. Haplogroup C is a founding lineage of the indigenous Amerindian, the seventh and youngest son of Japheth, Tiras.
Haplogroup mtDNA D is the principal East Asian lineage, with D1 found amongst Amerindians and D4 prevalent in Central Asians and much of Siberia, the descendants of Japheth’s third son, Madai.
Haplogroup E is found in Southeast Asia: in Malaysia, Borneo in Indonesia, the Philippines, Taiwanese aborigines and in Papua New Guinea – who are all descendants of Japheth’s fourth son, Javan.
Widespread Haplogroup G includes northeast Siberians, northern East Asians and Central Asians.
Haplogroup Q is found in Melanesian, Polynesian and New Guinean populations in southeast Asia and the Pacific, primarily descended from Noah’s grandson Javan.
The next major split is found in super Haplogroup N and its mutations are more widely spread globally than Haplogroup M.
Haplogroup A is found predominantly in many Amerindians and some East Asians and Siberians.
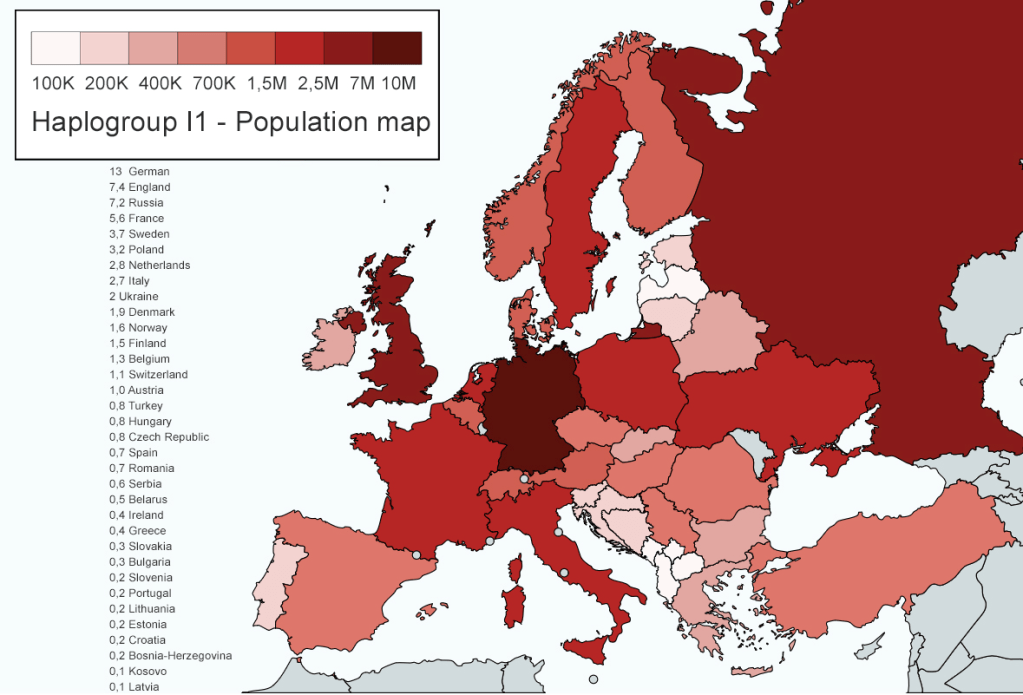
Haplogroup mtDNA I is the first mutation associated with Europeans, who are descended from Noah’s youngest son Shem (though may well originate with Ham’s line and his wife Na’eltama’uk). Haplogroup I is quite rare and ‘found in average in 2% of Europeans and under 1% of Near Easterners… Elevated concentrations are found in Daghestan’, for example Chechens (6%) and in isolated areas of Europe, such as Latvia (4.5%), Brittany (3%), Great Britain (4%), Ireland (3%) and Croatia (3%) to name a few. Haplogroup I is absent from the Basque country, which alternatively has high levels of mtDNA U5 and HV0+V. Haplogroup I sub-clades include I1 to I7 found in Armenia and Kuwait. It is thought to have been brought to Europe across the Caucasus.
Haplogroup S is specific to some Australian aborigines.
Haplogroup W is a crossover mtDNA Haplogroup in that it is commonly found in Eastern Europeans, as well as Central Asia, East Asia and southwest Asia. This means it is found in all three of Noah’s sons and daughters descendants. Maximum frequencies of W are observed in countries such as Finland (9.5%) and Hungary (5%). This is interesting as Finnish men (from Shem) exhibit high levels of Y-DNA Haplogroup N1c1 from admixture with Japheth. Finland shares with Hungary the unique and minority Finno-Ugric language group.
‘In Asia, haplogroup W is most common among the northern Pakistani… but is also found around 1.5% among the… Kazakhs, and at trace frequencies (< 0.5%) among many North Asian ethnic groups… Haplogroup W is descended from haplogroup N2.’
Geographic distribution of Haplogroup W matches the historical population movements of Y-DNA R1a for the Balto-Slavic speaking peoples. Haplogroup W is ‘considerably more common among the upper castes’ of India. Blond hair is believed to have originated within the R1a branch of the Indo-Europeans and therefore ‘propagated by women belonging’ to Haplogroup W. The Haplogroup like I is split into seven sub-clades. And like Haplogroup I, may well have a Hamitic origin or be the result of repeated admixture between the lines of Shem and Ham.
Haplogroup X is another crossover Haplogroup found in southern Siberians, Southwest Asians, North Africans and in Southern Europeans. ‘Haplogroup X is one of [the] rarest matrilinear haplogroups in Europe, being found only in about 1% of the overall population. The highest incidence of haplogroup X is observed in Greece [4%]… In Western Europe, X peaks in Orkney [7%], Scotland [2.5%], Catalonia 2.5%) and the Basque country [2.5%]. The only Eurasian ethnic group possessing a relatively high percentage of haplogroup X are the Druzes of Lebanon, Syria and Israel, among whom X makes up 15% of maternal lineages. The Druzes also have the greatest diversity of X lineages of any population…’ Its subgroup X2a is one of the founding lineages of indigenous North Americans; notably among the Sioux (15%).
Haplogroup X with I and W is one of the few ‘West Eurasian’ groups that does not descend directly from R but from the older macro-Haplogroup N, which is upstream of R. These are called ‘Basal Eurasian’ as they are closer to the N Haplogroup in the phylogenetic tree. The sixteenth President of the United States, Abraham Lincoln (1809-1865) ‘belonged to the very rare Haplogroup X1c’ which has been found in the Levant amongst the Druzes and in Tunisia. ‘Isolated samples have been reported in Italy, Ireland and Norway.’
This again with Haplogroups I and W, points to either a Hamitic origin (specifically Na’eltama’uk) or repeated intermixing. The fact it is a founding lineage for the Sioux Indian, hints at a Hamitic infusion in the Japhetic Line of the Sioux. In the mind of this writer, it raises a question as to the accuracy of the phylogenetic mtDNA tree. The apparent blurring between M, N and R and the Haplogroups downstream of each of them paints a confusing picture of the mutational evolution for the four main divergent races and the twenty-one principal ethnicities in the world.
Haplogroup Y is exclusively associated with Japheth like mtDNA Haplogroup A. Found in Siberian populations and at low frequencies in Central Asian, Japanese, Korean and Austronesian peoples.
The most recent significant maternal Haplogroup mutation is R. It is deemed ‘ancient and complex’ and is a large group literally found all over the world. Haplogroup R derives not from M but N and has the most Haplogroup mutations. Populations contained in Haplogroup R are divided ‘geographically into West Eurasia and East Eurasia. Almost all European populations and a large number of Middle-Eastern population today are contained within this branch.’
The first is Haplogroup B which is a principal East Asian lineage found in varying percentages amongst the Chinese, Tibetans, Mongolians, Central Asians, Koreans, South Siberians, the Japanese and Austronesians. With Haplogroups A, C and D, B is also found in the Amerindian.
Haplogroup F is one of the primary mitochondrial lineages in East and Southeast Asia. Its greatest frequency and sequence diversity can be found among coastal Asian populations, especially Vietnam. Enigmatically, F is found at 8.3% on Hvar Island in Croatia.
R0 is not East Asian and is found in Arabia, Ethiopia and Somalia. Following R0 is HV, the parent of Haplogroups H and V and found in Europe, Western Asia and North Africa. It is the most successful maternal lineage and dominates western European lineages, with over ‘half of the European population and between 25% and 40% of the Near Eastern population’ descending ‘from a single common female progenitor.’ This aspect more than hints at an origin which Shem’s wife and admixture with Ham’s descendants.
Most Europeans belonging to the HV lineage descend from a branch that was renamed H. A secondary though sizeable European branch was called V. There are seventeen sub-clades which are neither classified as H or V, ranging from HV0+V to HV13.
Haplogroup HV is found between 4% to 9% in the Middle East, for instance in Iraq (9%). In Europe, HV is very rare in Finland, Scandinavia, the British Isles, the Netherlands, Germany, Switzerland and Austria – all descendants of Abraham or his brother Haran (apart from Finland). The highest percentages for Haplogroup HV in Europe are observed in Italy, such as Calabria (10%) and Tuscany (5%) as well as in Ukraine (3.5%) and Greece (3%). The distribution of mtDNA HV is particularly reminiscent of Y-DNA Haplogroup T. ‘Haplogroup HV is found as far south as Ethiopia and Somalia, which are also hotspots of Y-haplogroup T.’ This is an interesting link between HV and the paternal Haplogroup T of Hamitic origin.
While Haplogroup HV is frequent in the Middle East, Haplogroups HV0 and V are rare. HV0+V are found in less than 1% of the Middle Eastern population and is almost absent from the Arabian Peninsula. Haplogroup V has 21 sub-clades ranging from V1a1a in Scandinavia, Finland and the Baltic to V20 in Norway. Both Benjamin Franklin the American founding father and Bono from rock band U2 are mtDNA Haplogroup V.
Haplogroup H [2] is the most common mtDNA Haplogroup of all, as well as the most diverse maternal lineage throughout the northern Hemisphere. There are many basal sub-clades of Haplogroup H, including up to H95a. H1 for instance, is found from Europe and North Africa to ‘the Levant, Anatolia, the Caucasus, and as far as Central Asia and Siberia.’ The frequency of Haplogroup H in Europe ranges between 40% and 50%. The lowest frequencies are observed in for instance, Finland (36%) and Ukraine (39%). Regions where it exceeds 50% include Galicia (58%) in northern Spain and Wales (60%).
‘The Cambridge Reference Sequence (CRS), the human mitochondrial sequence to which all other sequences are compared, belongs to haplogroup H2a2a.’ Certain H sub-clades are ‘rare in Europe and geographically confined mostly to the Middle East. This includes H14 and H18.’ Though the precise sub-Haplogroup is unclear, the lineage of Queen Victoria belongs to mtDNA Haplogroup H. Napoleon Bonaparte possessed the rare 16184T mutation within Haplogroup H15a1b. In Europe, H15 is found in Scotland, Germany, Poland, Austria and northern Italy; while H15a is found mostly in northwestern Europe including Scotland.
The remaining mtDNA Haplogroups includes Pre-JT which splits into J and T. Haplogroup J [3] is one of four major European-specific Haplogroups and is evenly distributed across Europe. The highest frequencies of Haplogroup J include: Cornwall (20%), Wales (15%), Iceland (14%), Denmark (13.5%), Scotland (12.5%), England (11.5%), Switzerland (11.5%) and the Netherlands (11%). As Haplogroup HV is rare amongst the descendants of Abraham, Haplogroup J is relatively frequent (following Haplogroup H).
In the Middle East, it is most frequently found in countries such as Saudi Arabia (21%) and Iraq (13%). Haplogroup J is split into J1 and J2 with many sub-Haplogroups within each.
Haplogroup T [4] is one of the youngest Haplogroup mutations and is composed of two main branches T1 and T2. ‘The two of them have very different distributions, which are diametrically opposed in most regions.’ The highest percentages of T1 include the Udmurts (15%) of the Volga-Ural region of Russia, Romania (6%) and Iraq (5.5%). While Haplogroup T2 also peaks among the Udmurts (24%) and is frequently encountered in the Netherlands (12%). Haplogroup T2b is of interest to this writer and is found in higher percentages in Europe, especially around the Alps and is commonly found in Britain (T2b4b, T2b4d, T2b4f), particularly in England (T2b2b, T2b19, T2b24) – as well as in Scotland, (T2b9) and Ireland, (T2b13).
Haplogroup U [1] is one of Europe’s oldest and most diverse Haplogroups, with numerous sub-clades. About 10 to 11% of Europeans and European Americans belong to U.
Haplogroup U5 is prevalent in Europe, between 5% to 12% and in particular shows high frequency in Scandinavia and the Baltic countries with the highest percentage in the Sami people. Outside of Europe, U exhibits a high frequency in the Indian sub-continent (U2, U7) and in North Africa, where U6 is common.
Finally, there is Haplogroup K, where certain lineages are found in Central Asia and Northern Africa. In Europe, mtDNA K is common in northwest Europe, peaking in Belgium (14%), then Ireland (12%), the Netherlands (10%), Iceland (10%), Denmark (9%) and France (8.5%). As with Haplogroup J, mtDNA K is prevalent amongst Abraham’s descendants.
‘In the Eastern Mediterranean and the Middle East, haplogroup K reaches high frequencies in Cyprus (20%), among the Druzes of Lebanon (13%), [and] in Georgia (12%)…’ K1a1a for instance is found in Central Asia, as well as central and western Europe and could be linked to the diffusion of R1b. K1c is common in Central Asia; while K1c1 is common in Slavic countries. K1c2 is more common in Germanic countries. Both could be associated with R1a.
Haplogroup K ‘is known for its presence in distinct population groups, such as the prehistoric Basques and the Ashkenazi Jews. Ashkenazi Jews are the ethnic group with the highest percentage of mtDNA K lineages today: 32% in average, and up to 50% among Ashkenazi Jews from Germany. There are only three typically Jewish subclades of K: K1a1b1a, K1a9, and K2a2a. There are other subclades, like K1a7, K1a8 and K2c, which are also found among people of Jewish descent, but they are very rare.’
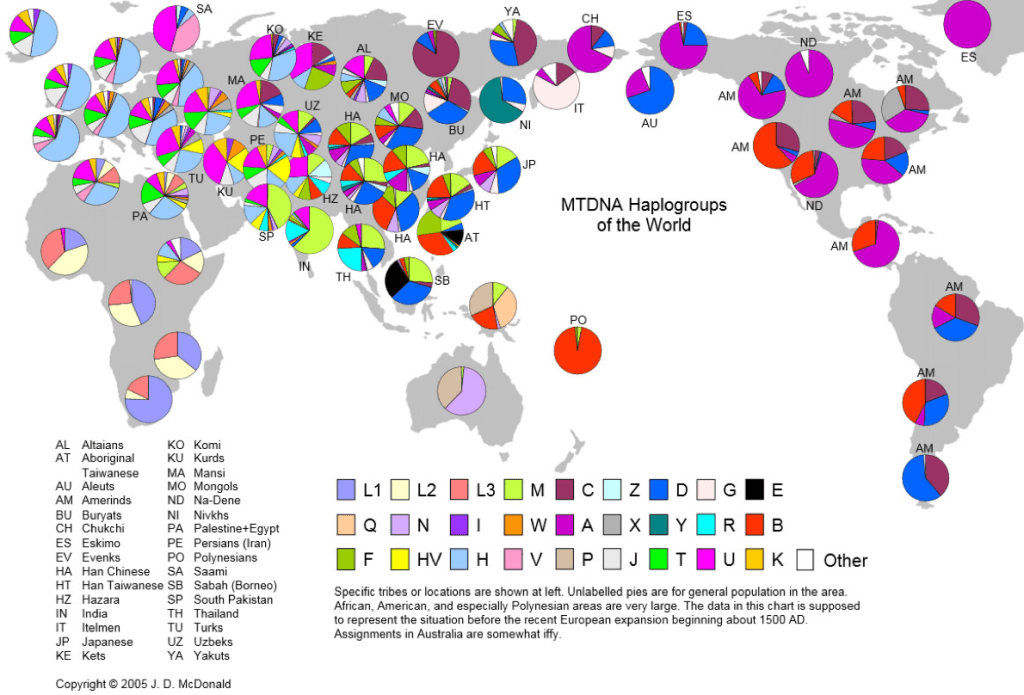
Analysing the Haplogroup family trees of the world, it is evident, that mtDNA passed from mothers to all their children, includes (alphabetically) the principal Haplogroups for ‘Adataneses (Japheth) of A, B and D; the key Haplogroups for Na’eltama’uk (Ham) of H, M and U; and (for the Canaanites) Haplogroup L; while the main Haplogroups for Sedeqetelebab (Shem) are H, U, J, T and K. Notice the crossover Haplogroups H and U, which each logically contain more diverse mutations and are further widespread, than any other maternal mtDNA Haplogroup.
DNA from the Y chromosome passed from fathers to their sons is perhaps a more reliable and stable tracker for lines of lineal descent and lettered as follows.
A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, P, Q, R, S and T.
African:
A, B, E1b1a
West Eurasian – including North Africa, the Middle East and South Asia:
E1b1b, G, H, I1, I2, J1, J2, L, T, R1a, R1b, R2
East Eurasian and Native American:
C, D, K, N, O1, O2, Q
Austronesian-Melanesian:
M, S

The male Y sex chromosome Haplogroups are divided broadly into four major groups on the phylogenetic tree which in turn produce a sequence of different Haplogroup mutations.
The first is Haplogroup BT = C, D, E & (F); then F = G, H, I, J & (K); K = L, T, N, O, M, S & (P); and Haplogroup P = Q, R.
The most ancient Y-DNA Haplogroup originating from Y-DNA Adam, is A. Like Haplogroup B, it only appears in Africa. Haplogroup A is indicative of sub-Saharan Africans and the oldest clade of A00a, L1149, known as ‘Perry’s Y’ was discovered in 2012 in an African American. Haplogroup A00 was first discovered in Mbo Bantu men from West Cameroon. Bantu can vary in skin tone from light brown to medium brown. The highest concentration of Haplogroup A00 found in 2015, belonged to the Bangwa Grassfields Mbo Bantu.
‘Haplogroup A is the NRY (non-recombining Y) macrohaplogroup from which all modern paternal haplogroups descend. It is sparsely distributed in Africa, being concentrated among Khoisan, M91 populations in the southwest…’ Recall the Khoisan also possess the ancient mtDNA Haplogroup L0. Haplogroup B, M60 is prevalent amongst the Pygmies in Africa. Both A and B are without the M168 (and M294) mutation that defines all other Haplogroups, beginning with C, D and E. ‘Haplogroup BT is a sub-clade of Haplogroup A, more precisely of the A1b clade…’
- Haplogroup A
- Haplogroup A00 (F6)
- Haplogroup A0 (formerly also A1b) (V148)
- Haplogroup A1 (also A1a-T)
- Haplogroup A1a (M31)
- Haplogroup A1b (also A2-T; P108, V221)
- Haplogroup A1b1a1 (also A2; M14)
- Haplogroup A1b1b (also A3; M32)
- Haplogroup BT (M91, M42, M94, M139, M299)
- Haplogroup B (M60)
- Haplogroup CT (P143)
It is fair to say that Adam as Y-DNA Adam, possessed Haplogroup A and specifically A00. For this to transfer to the post-diluvian age, Noah must have carried the same Haplogroup, passing either A00 or early mutations of A0 and A1 to his three sons. Subsequent mutations likely began with Noah’s twenty-one grandsons.
What is of note, is that it is Noah’s illegitimate fourth son Canaan’s male descendants, who carry Haplogroups A and B – refer Chapter XI Ham Aequator.
Haplogroup C (M130, M216) is the first clade which is not indicative of sub-Saharan African men. Though C1 is found in low frequencies in India, C1b1a1 (M356); Europe, C1a2 (V20); and the Australian Aborigine, C1b3b (M347); it is almost exclusively a defining marker Haplogroup for the descendants of Japheth. For instance, C1a1 (M8) in Japan; C1b1a2b (F725) in China; C1b3a (M38), indicative of males in Indonesia, New Guinea, Melanesia, Micronesia and Polynesia; C2, (M217) common amongst Mongols and the predominant Haplogroup for the Kazakhs of Central Asia at 40%; and P39 in the Amerindian.
Haplogroup D with E derives from its parent clade DE (M1, M145, M203). D1 (M17) is found exclusively amongst Japheth’s descendants. D1a1 (M15, P99) is found in Tibetan men (52%) and D1a2 (M55) is found in Japanese men at 40%. Whereas D2 is peculiar to Nigeria, Saudi Arabia, Syria and African American men and thus perhaps misnamed as Haplogroup D
Haplogroup E (M40, M96) on the other hand could not be more different from its supposed mutational relative Haplogroup D. Thus far, Haplogroup A and Haplogroup B are exclusively sub-Saharan (with the exception of M13 [A1b1b2b]) and C with D are almost entirely East Asian. Whereas Haplogroup E is very much split between its origin in sub-Saharan Africa with those men somewhat related in North Africa and the Middle East on one hand; and those peoples with Haplogroup E predominantly located in the Balkans, yet also scattered throughout the majority of European nations.
Countries with high percentages in their male populations include Montenegro, 27%; Macedonia, 21.5% and Greece, 21%. The former two have more men with Haplogroup I2a1 and in Greece there are more with J2. The Island of Sicily has 20% and ‘Ashkenazi [Jewish men] also exhibit approximately 20% of E1b1b, which falls mostly under specific clades of E-M123 [E1b1b1b2a1].’
European nations with E1b1b as the majority male Haplogroup include, Kosovo, 47.5%; Albania, 27.5% and Bulgaria, 23.5%. Most universally assume that the strain V13 is evidence of a bona-fide European lineage of E1b1b. Yet, the contention exists that it more accurately proves a mutated lineal descent from either African males who originally possessed E1b1b (M215); or Arab related peoples who carry E1b1b from admixture themselves.

Haplogroup E is one of the most branched groups – in parallel with the descendants of Canaan, represented by sub-Saharan Africans (Chapter XII Canaan & Africa) – with many sub-Haplogroups. Haplogroup E mutated into E1 and E2 (M75) which is found in sub-Saharan Africans, for instance the Zulu with 20.69%. E1 split into E1a (M33, M132]) formerly E1 and E1b (P177) formerly E2.

From E1b derives E1b1 formerly E3 and then again into E1b1a (V38) an ancient brother to E1b1b, but which has left a completely different fingerprint on the world today. Haplogroup E1b1a is as indicative of Black Africans from Canaan as the mutated E1b1b is for the Berbers who are a mixture with Ham’s son Mizra. In fact, E1b1a formerly E3a, is the defining marker Haplogroup for African males. Far more predominant than either Haplogroup A and B. E1b1a diversified into E1b1a1 (M2) – the Niger-Congo speaking peoples, the most common and diversified Haplogroup in West Africa between 70 to 97% – and E1b1a2 (M329) found in Ethiopia and Omotic speaking populations. Haplogroup E1b1b mutated into E1b1b1 (M35), found in the Horn of Africa, North Africa, the Middle East, the Mediterranean and the Balkans.
All these peoples share mutual paternal ancestors and so the Mediterranean and Balkan males, for instance the Greeks with E1b1b (formerly E3b), are in one sense more related to the Berbers of North Africa who they share E1b1b than they are to other Greek men say, who carry J2, R1b, or R1a Haplogroups. Therefore if accurate, the parent clade DE highlights a partial blurring between Black African and East Asian genes and thus reveals Black people and East Asians could have more in common paternally than they do with Europeans.
Support for this may lay in the fact that both Ham’s wife Na’eltama’uk and Japheth’s wife ‘Adataneses were of different descent from Adam and Eve’s son Seth of whom Noah (and probably his wife) descended. For Na’eltama’uk was of the line of Cain; who’s mother was Eve, but Cain’s father was not Adam – refer Chapter XXII Alpha & Omega. ‘Adataneses was from the Neanderthal line of Day Six of creation and so she was not related to Noah in part, as Na’eltama’uk or fully, as Shem’s wife Sedeqetelebab – Chapter XXII Alpha & Omega.
From an early humankind perspective the shared Japheth-Ham DE clade is in contrast to the F mutation from which Europeans descended from Shem appear to wholly derive.
Aside from Kosovo with 47.5% E1b1b, other nations with high percentages include: ‘Morocco (over 80%), Somalia (80%), Ethiopia (40% to 80%), Tunisia (70%), Algeria (60%), Egypt (40%), Jordan (25%), Palestine (20%), and Lebanon (17.5%).’
All this data reveals beyond question a Canaanite origin and infusion of Haplogroup E(1b1b) primarily into Hamitic lines and to a lesser degree into Shem’s male descendants.
Famous persons of note perhaps considered fully European, yet their Y-DNA Haplogroup E1b1b descent saying otherwise, includes:
Skanderbeg (Albanian feudal lord); Giuseppe Garibaldi; Lyndon B Johnson (36th President); Napoleon; Albert Einstein; Nicolas Cage; Franz Kafka; Caravaggio (baroque painter); Adolf Hitler; Zinedine Zidane; the Wright brothers; Clan Colquhoun (Calhoun); Larry Page (Google co-founder); William Harvey (blood circulation); Steven Pinker (psychologist/scientist); David Attenborough (broadcaster); Richard Attenborough (film director); Tom Conti (actor).
Perhaps surprise inclusions include Nelson Mandela, Desmond Tutu and Ramesses III, because the first two men at least, one would have thought they were Y-DNA Haplogroup E1b1a (or E1a, E2, A, B) instead.

For example the men above all look ostensibly white (European), yet a closer inspection of their physiognomy indicates the plausibility for a black paternal ancestor, resulting in not just distinctive faces – such as born by Adolf Hitler, Nicholas Cage and Larry Page – but exhibiting mixed facial features akin to someone from North Africa (Berber) or the Middle East (Arab).
The next Haplogroup after the intriguing E mutation, is group F. The Haplogroups descending from macro-Haplogroup F are found in some 90% of the world’s male population and almost exclusively outside of sub-Saharan Africa. F is the immediate parent of Haplogroups G, H, I, J and K, a further macro-Haplogroup. However, excluding these common Haplogroups the sub-clade F* (M89) – and F1 (Sri Lanka), F3 (M48), India and Nepal] – appears in the Indian sub-Continent countries of India and Pakistan, peaking in Sri Lankan males with 10%.
F2 (M427) on the other hand is found in minorities located in Southern China and Continental South East Asia. Rather like D, Haplogroup F is split between mainly Cush and Phut from Ham and partially East Asians from Japheth. F1 (P91), F2 and F3 (formerly F5), are all quite rare and exclusive to the regions where they are located. ‘In such cases, however, the possibility of misidentification is considered to be relatively high and some may belong to misidentified subclades of Haplogroup GHIJK.’ Haplogroup FT (P14, M213) also has the M89 mutation and is found in China, Vietnam and Singapore.
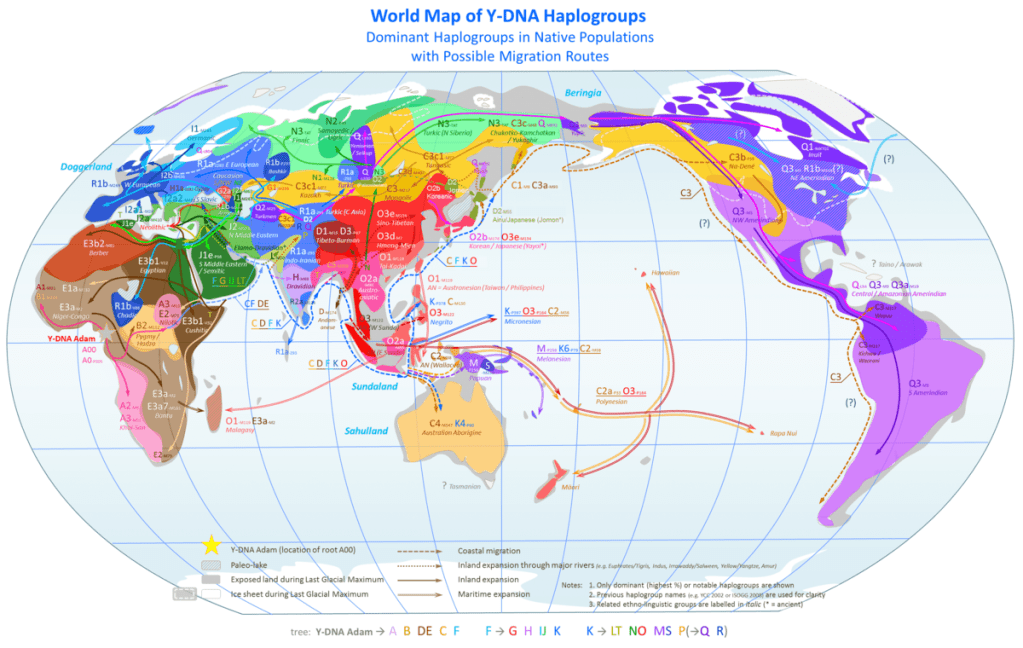
The first Haplogroup mutation from the major ancestor intersection group of F is Haplogroup G. It is an ancient lineage, though unlike Haplogroups A through to E which preceded it, its paternal origin and lineage is not as clearly delineated. Though undoubtedly it is the first identifiable Haplogroup for Shem’s descendants. It is probably an early precursor lineage to Haplogroup I which is similarly spread throughout many nations, but even when a majority frequency, it isn’t a defining marker Haplogroup for the peoples of that country – with the exception of Georgia.
An online encyclopaedia states: ‘In 2012, a paper by Siiri Rootsi et al. suggested that: “We estimate that the geographic origin of haplogroup G plausibly locates somewhere nearby eastern Anatolia, Armenia or western Iran. Previously the NGS placed its origins in the Middle East 30,000 years ago and presumes that people carrying the haplogroup took part in the spread of the Neolithic.
Two scholarly papers have also suggested an origin in the Middle East, while differing on the date. Semino et al. (2000) suggested 17,000 years ago. Cinnioglu et al. (2004) suggested the mutation took place only 9,500 years ago. A more eastern origin has also been mentioned, believed by some to originate in an area close to the Himalayan foothills.’ Two important points are highlighted here. First, the time scale suggesting 9,500 years ago is supported by an unconventional chronology. Second, an origin at the Himalayan foothills concurs with this writer’s research – refer Chapter I Noah Antecessor Nulla.
Haplogroup G (M201) splits into two main divisions, G1 (M285, M342) and G2 (P287). Haplogroup G2 is more prolific and divides into G2a (P15) and G2b (M377). Haplogroup G is found in Western Europe, Northwest Africa, East Africa, Central Asia and India. Even so, it is a minority male Haplogroup with frequencies of between 1 and 10% of the population. The exceptions are the Caucasus region and parts of central, southern Italy and Sardinia, where frequencies range from 10% to 30% of male lineages. The highest percentages are found in Georgians (30%), Karachay-Cherkessians (40.5%), Abkhazians (47.5%), the Adygei (53.5%) and Ossetians (56%). ‘The highest genetic diversity within haplogroup G is found in the northern part of the Fertile Crescent, between the Levant and the Caucasus…’
European men in the main who exhibit Haplogroup G belong to the G2a sub-clade; with most northern Europeans where it is relatively rare and Mediterranean Europeans primarily within either L140 (G2a2b2a1) or M406 (G2a3a). Almost all carriers of G2b (L72+, formerly G2c [G5]) found in Europe are Ashkenazi Jews, G2b is found in the Middle East and Pakistan. Similarly, Haplogroup G1 is found predominantly in Iran and also in the Levant among Ashkenazi Jews, as well as in Central Asia, primarily in Kazakhstan.
G2a is generally located in the mountainous regions of Europe, whereas ‘some sub-clades of L140 are found uniformly throughout Europe, like Scandinavia and Russia…’ as well as ‘the Caucasus, Central Asia and throughout India, especially among the upper castes, who represent the descendants of the Bronze Age Indo-European invaders. The combined presence of G2a-L140 across Europe and India is a very strong argument in favour of an Indo-European dispersal… [where] G2a-L140 came from Anatolia to eastern and Central Europe… (a fact proven by ancient DNA test). Once in Southeast Europe men belonging to the U1 [G2a3b1a1] branch founded the Cucuteni-Trypillian culture (with men of other haplogroups, notably I2a1b-L621 around modern Moldova). The oldest known G-L293 [G2a1] sample is a Neolithic man from western Iran. Nowadays, G-L293 is the most common G2a clade in the central and northern Caucasus, peaking at 64% of the population in North Ossetia.’
This concentration Of G2a in West Asia, comprising the Caucasus (Azerbaijan, 18%; Armenia, 11%), Turkey (11%) and Iran (10%+) lends support to two conclusions.
First, the origin of Haplogroup G like I1 and I2 is associated with Shem and not Ham. Support for this is that in sub-Saharan Africa, G is rarely found among native populations. In the Middle East, it accounts for only about 3% of the population in almost all areas, including North African Berbers. About 10% of Jewish males are Haplogroup G.
Second, as with Haplogroup I or E1b1b from Canaan, it is an ancient mutation found in a minority of Shem’s descendants, scattered within countries where other Haplogroups are the defining marker, such as R1a and R1b. Higher than average percentages for G exist in various parts of Eastern Europe, like I2a1. For instance: ‘In the Tirol (Tyrol) of western Austria, the percentage of G-M201 can reach 40% or more… In north-eastern Croatia, in the town of Osijek, G was found in 14% of the males. Farther north, 8% of ethnic Hungarian males and 5.1% of ethnic Bohemian (Czech) males have been found to belong to Haplogroup G’ and ‘In Wales, a distinctive G2a3b1 type (DYS388=13 and DYS594=11) dominates there and pushes the G percentage of the population higher than in England.’
Encyclopaedia: ‘Three of the main maternal lineages thought to have evolved conjointly with Y-haplogroup G2 are mt-haplogroups N1a1a, W1 and X, all minor lineages… Interestingly, N1a, W (aka N2b) and X are directly descended from the very old haplogroup N*, rather than from the more recent macro-haplogroup R (the ancestor of HV, JT and UK, representing 90% of European mtDNA lineages). The long bottleneck evolution of N1a and X mirror that of Y-haplogroup G2. These haplogroups are called Basal Eurasian.’
Famous men included in carrying Haplogroup G: Joseph Stalin, G2a1a (originally from Georgia); Al Capone, G2a-P303 (G2a2b2a); Larry Bird G-Z6748 (American Basketball player); and Jewish actor Jake Gyllenhaal.
Haplogroup H as with G, shares the same M89 mutation stemming from Haplogroup F. Haplogroup H is a lineage from Noah’s son Ham and descends primarily through his son, Cush. It is prevalent in the Indian sub-Continent in the form of H1 and the rarer H3. ‘Its sub-clades are also found in lower frequencies in Iran, Central Asia, across the middle-east, and the Arabian peninsula.’ H2 (P96) formerly F3, is present in Europe and western Asia.
The principal sub-Haplogroups for Y-DNA Haplogroup H.
H-L901/M2939 is a direct descendant of Haplogroup GHIJK. There are three direct descendants and their defining SNPs are as follows:
- H1 (L902/M3061)
- H1a (M69, M370)
- H1b B108, Z34961, Z34962, Z34963, Z34964
- H2 (P96, L279, L281, L284, L285, L286, M282)
- H2a FGC29299/Z19067
- H2b Z41290
- H2c Y21618, Z19080
- H3 (Z5857)
- H3a (Z5866)
- H3b (Z13871)
The primary branch of H1 is the most predominant Haplogroup (H1a) amongst populations in South Asia particularly its descendant H1a1* (M52). A branch of M52, H1a1a (M82), is commonly found among the Romani in the Balkans (60%) who originated in South Asia, migrating into the Middle East and Europe, from the beginning of the second millennium CE and the Medieval period. H1a (M69) is common amongst populations living in Bangladesh, India, Sri Lanka and Nepal; while in the Pashtuns of Afghanistan (6.1%) and Pakistan (4.2%) it is not as common.
The highest percentages of H1a are found in Dravidian men of southern India with 32.9%; Bangladesh at 35.71%; and Sri Lanka with 25.3%. In northern India, Haplogroup H is most commonly found amongst Rajput men at 44.4%. Haplogroup H1a is found in Europe, Central Asia and South East Asia, though in very small percentages as evidence of admixture and intermarrying.
Haplogroup H1b was only discovered in 2015. It was ‘detected in a single sample from an individual in Myanmar. Due to only being classified recently, there are currently no studies recording H1b in modern populations.’ Haplogroup H2 is the only primary branch of H located mainly outside South Asia. Known as F3, H2 was reclassified as Haplogroup H as it shared the marker M3035 with H1. H2 has been found in a number of ancient samples, yet only rarely in ‘modern populations across West Eurasia.’
H2 is commonly found with G2a samples, with two main clades of H2m and H2d. ‘H2d was found along the inland/Danubian route into central Europe, but most H2m individuals are found along the Mediterranean route into Western Europe, the Iberian Peninsula and ultimately, [in] Ireland. There were also two occurrences of H2a found in the Neolithic Linkardstown burials in the southeast [of] Ireland. More Neolithic H2 samples have been found in Germany and France. H3 like H2 is newly classified and is not readily found in modern population studies. Samples belonging to H3 have been labeled under F*. In consumer testing, it has been found principally among South Indians and Sri Lankans, and other areas of Asia such as’ in Bahrain and Qatar.
Haplogroup I (M170) is a clear European paternal Haplogroup and considered the oldest major Haplogroup in Europe. Yet its roots likely lay in the earlier G Haplogroup. Haplogroup I ranges from frequent to infrequent in European males and though spread across Europe it is principally found in two distinct locations resulting from a mutational split. I-M170 is not part of the M89 mutation which bonds Haplogroups F, G and H. It derives originally from Haplogroup IJK, L15/L16 mutations and then IJ (M429). Haplogroup I is found sporadically in the Middle East due to admixture and is virtually absent elsewhere in the world.
Encyclopaedia: ‘Haplogroup I appears to have arisen in Europe, so far being found in Palaeolithic sites throughout Europe (Fu 2016), but not outside it. It diverged from common ancestor IJ*… (Karafet 2008).’ This writer is not convinced* this is necessarily the case; as Haplogroups I1 and I2 are paternal marker Haplogroups for Shem’s descendants, while J1 and J2 are related to Ham.
‘Early evidence for haplogroup J has been found in the Caucasus and Iran (Jones 2015, Fu 2016). In addition, living examples of the precursor Haplogroup IJ* have been found only in Iran, among the Mazandarani and ethnic Persians from Fars. This may indicate that IJ originated in South West Asia. Haplogroup I has been found in multiple individuals belonging to the Gravettian culture… [which] expanded westwards from the far corner of Eastern Europe, likely Russia, to Central Europe. They are associated with a genetic cluster that is normally called the Věstonice cluster.
The earliest documentation of I1 is from Neolithic Hungary, although it must have separated from I2 at an earlier point in time. In one instance, haplogroup I was found far from Europe, among 2,000-year-old remains from Mongolia. The role of the Balkans as a long-standing corridor to Europe from Anatolia and/or the Caucasus is shown by the common phylogenetic origins of both haplogroups I and J in the parent haplogroup IJ (M429). I and J were subsequently distributed in Asia and Europe in a disjunctive phylogeographic pattern typical of “sibling”* haplogroups. The existence of Haplogroup IJK – the ancestor of both haplogroups IJ and K (M9) – and its evolutionary distance from other subclades of Haplogroup F (M89), supports the inference that haplogroups IJ and K both arose in Southwestern Asia. Living carriers of F* and IJ* have been reported from the Iranian Plateau.’

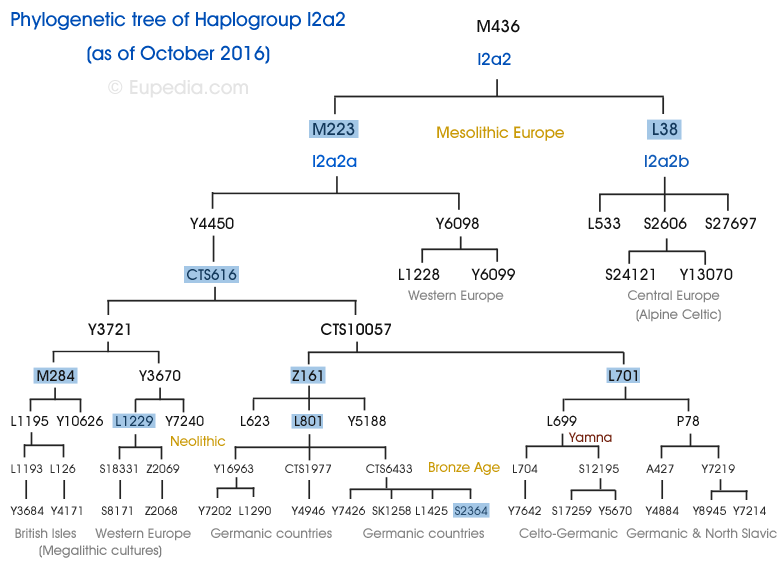
Haplogroup I split into the key Haplogroup divisions of I1 (M253) and I2 (M438). Haplogroup I1 is dominant in Nordid and Nordic Europids of Scandinavia and north western Europe; whereas I2 is located primarily in Dinarid and Dinaric Europids of Central and southeast Europe, Sardinia and the Balkans. There is one mutation of I2 which enigmatically occurs more frequently in northwest Europe – M223. Though since 2018 I2a2 is now known as I2a1b1. The main mutations shown on the map above include: I1 (M253); I2a1 (P37.2); I2a1a (M26); I2a1b (M423); and I2a2a [I2a1b1] (M223).
The main I Haplogroups consist of the following classifications:
I M170
I1 M253
I1a DF29
I1a1 CTS6364 / Z2336
I1a2 Z58
I1a3 Z63
I1b Z131
I1c Z17925
I1d Y19086
I2 M438
I2a L460
I2a1 P37.2
I2a1a M26
I2a1b M423
I2a2 M436
I2a2a [I2a1b1] M223
I2b L415
I2c L596
Haplogroup I1 is found mostly in Scandinavia and Finland, where it typically represents Y chromosomes of 35% of men. I1 is associated with the Norse ethnicity and is found in regions invaded by the Vikings and ancient Germanic tribes. ‘After the core of ancient Germanic civilisation in Scandinavia, the highest frequencies of I1 are observed in other Germanic-speaking regions, such as Germany, Austria, the Low Countries, England and the Scottish Lowlands’ which all have I1 lineages averaging between 10% to 20%.’
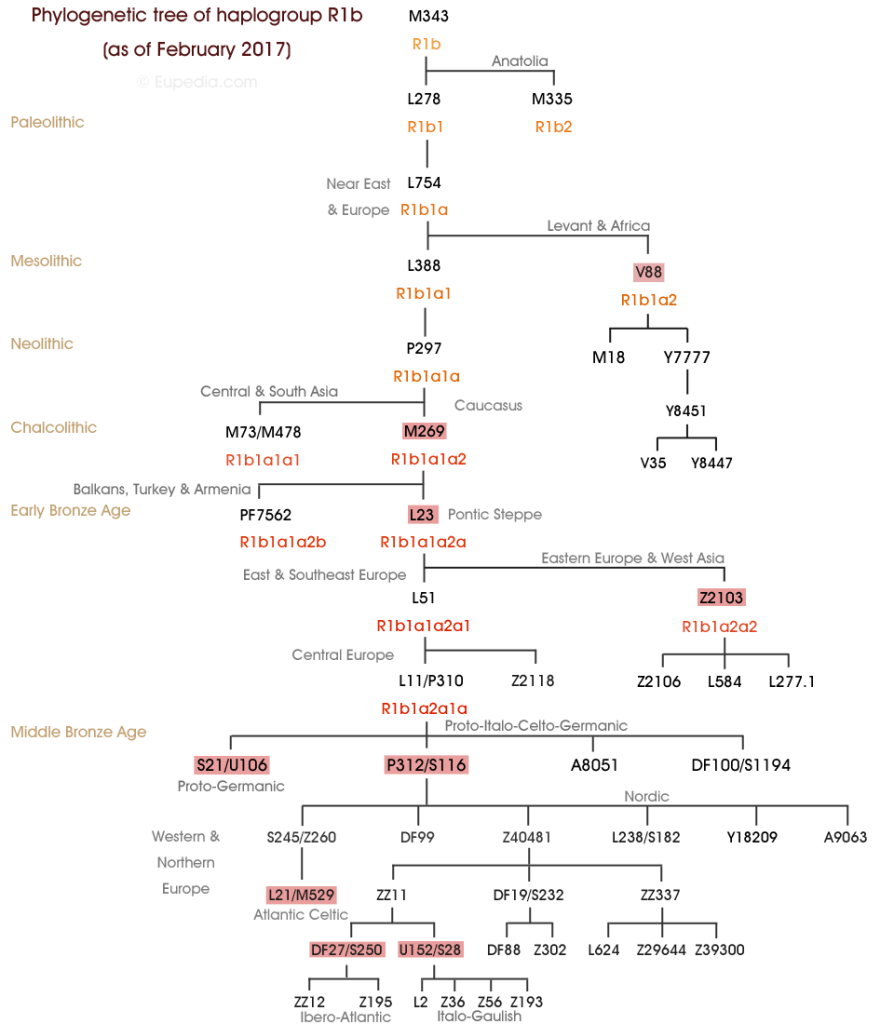
Thus, as we saw with mtDNA Haplogroups J and K, Y-DNA Haplogroup I1 is the most frequently present in nations containing the offspring of Abraham. Yet what is vital to appreciate is that Haplogroup I1 is indicative of a lineage from Peleg and older than the R1b-U106 predominantly carried by Abraham’s male descendants – Chapter XXVII Abraham & Keturah – Benelux & Scandinavia.
Encyclopaedia: ‘Outside Fennoscandia, distribution of Haplogroup I1-M253 is closely correlated with that of Haplogroup I2a2-M436; but among Scandinavians (including both Germanic and Uralic peoples of the region) nearly all the Haplogroup I-M170 Y-chromosomes are I1-M253.’

‘Another characteristic of the Scandinavian I1-M253 Y-chromosomes is their rather low haplotype diversity (STR diversity): a greater variety of Haplogroup I1-M253 Y-chromosomes has been found among the French and Italians, despite the much lower overall frequency of Haplogroup I1-M253 among the modern French and Italian populations. This, along with the structure of the phylogenetic tree of I1-M253 strongly suggests that most living I1 males are the descendants of an initially small group of reproductively successful men who lived in Scandinavia during the Nordic Bronze Age.’

‘L22+ (aka S142+) is a… big Nordic branch. It is… very common in Britain, especially on the east coast where the Vikings settled most heavily, in the Low Countries and Normandy… [the heritage of the Danish Viking], as well as in Poland and Russia (Swedish Vikings). Z58+ is chiefly West Germanic, with a… strong presence in Germany, the Low Countries and Britain. It is… found to a lower extent in Nordic countries and throughout Continental Europe. Its age has been estimated around 4,600 years before present. Z138+ (aka Z139+) is a… disparate subclade. It is found at very low frequency throughout the Germanic world, with a peak in England and Wales… it has also been found in Ireland, Portugal, southern Italy, Hungary and Romania. Z60+ is found throughout the Germanic world. Z63+ is a strongly Continental Germanic subclade, virtually absent from Nordic countries. It is most common in Central Germany, the Benelux, England, Lowland Scotland, as well as Poland.’
Haplogroup I2, M438 is the most common paternal lineage in former Yugoslavia, Romania, Bulgaria as well as in Sardinia. It is a lineage (I2a1b, M423) in many Slavic countries. ‘Its maximum frequencies are observed in Bosnia (55%, including 71% in Bosnian Croats), Sardinia (39.5%), Croatia (38%), Serbia (33%), Montenegro (31%), Romania (28%), Moldova (24%), Macedonia (24%), Slovenia (22%), Bulgaria (22%), Belarus (18.5%), Hungary (18%), Slovakia (17.5%), Ukraine (13.5%), and Albania (13.5%).’

Today, ‘I2a1, P37.2 is five to ten times more common than G2a in Southeast Europe, while during the Neolithic period G2a was approximately four times more common. What can explain this complete reversal?’ A possible answer is due to the fact that as the defining marker Haplogroups R1a and R1b for Europeans are voluminous in their dispersion compared with Haplogroup I, from which they descend; so to is Haplogroup I compared with the older Haplogroup G from which it in turn descends.

An interesting I2 sub-clade is I2a1a-M26. It is notable for its strong presence in Sardinia, where it dominates comprising 40% of Haplogroup I patrilineal lineages. Haplogroup M26 is virtually absent east of France and Italy. It is found in low frequencies in the Balearic Islands, Castile-Leon, the Pyrenees, southern and western France, parts of the Maghreb in North Africa, Great Britain, Ireland and the Basque Country. It is the only sub-clade of I-M170 found among the Basques. Fascinatingly, the M26 mutation is found in indigenous males inhabiting every ‘geographic region where megaliths may be found, including such far-flung and culturally disconnected regions as the Canary Islands… Corsica… and Sweden.’

The distribution of Haplogroup I2a2, M436 and I2a2a (I2a1b1), M223 closely correlates with that of Haplogroup I1 except in Scandinavia and Finland. It is thought that the lack of correlation between the distributions of I1 and I2a2 in Fennoscandia is a result of Haplogroup I2a2 being affected in the ‘earliest settlement of this region by founder effects and genetic drift due to its rarity.’ A sub-clade of Haplogroup I2a2, namely I2a2a1, M284 is found almost exclusively among the population of Great Britain. This indicates that the clade may have a long historical presence on the island. It is more than a coincidence that distribution of M253 and M436 correlate with the Germanic peoples historical migrations.
Both Haplogroups have been detected in Bithynia and Galatia in Turkey, areas linked with the ancient Gauls of Thrace, invited by Nicomedes I of Bithynia. ‘This suggestion is supported by recent genetic studies regarding Y-DNA Haplogroup I2b2-L38 [which] have concluded that there was some Late Iron Age migration of Celtic La Tène people, through Belgium, to the British Isles including north-east Ireland.’

There is an interesting link between height and Haplogroup I in Europe. Nations with taller than average men, such as the Netherlands, Scandinavia and in the Balkans all have higher than average Haplogroup I percentages in their male populations. The averages in the Dinaric Alps are reputed to be the tallest in the world, with an average male height between 180 cm (5 ft 11 in) to 182 cm (6 ft 0 in) in the cantons of Bosnia; 184 cm in Sarajevo; and 182 cm to 186 cm (6 ft 1 in) in the cantons of Herzegovina.
‘A 2014 study examining the correlation between Y-DNA haplogroups and height found a correlation between the haplogroups I1, R1b-U106, I2a1b and tall males. The study featured the measured average heights of young German, Swedish, Dutch, Danish, Serbian and Bosnian men. The German male average height was 180.2 cm, the Swedish men were on average 181.4 cm, the Dutch men were 183.8 cm, the Danish men were 180.6 cm, the Serbians were 180.9 cm, and men from Herzegovina were 185.2 centimeters on average.’
Famous male members of Haplogroup I1 include: Clan Hamilton, Z63; Clan Lyon, L22; Richard Henry Lee, founding father, L22 and his descendant Robert E Lee, Commander of the Confederate States Army during the Civil War; James Wilson, founding father; Alexander Hamilton, founding father, Z58; Andrew Jackson, 7th President; Ludwig van Beethoven, I1a Z138 from Z58; Samuel Morse, inventor and painter, L22; Leo Tolstoy; Chester A Arthur, 21st president, Z63; John Harvey Kellog Z58; Calvin Coolidge, 30th President; William Faulkner, Z60; Chris Pine, actor, I1-A13819; Robert I of Scotland, Clan Bruce, I1-Y17395; Jimmy Carter, 39th President; Warren Bufffet, business magnate and multi billionaire; Bill Clinton, 42nd President; Sting – Gordon Matthew Thomas Sumner.
Famous members from Haplogroup I2a1 include: Martin Luther, I2a, L147.2; Novak Djokovic, I2, PH908 downstream of L147.2; Clan Monroe, I2a1a, L161.1; Clan Lindsay I2a1a, L233; Clan Barclay, I2a1a, M26.
Famous members of Haplogroup I2a2 include: House of Clinton, I2a2a [I2a1b1], M223; George Clinton founding father and 4th Vice President; Bill Gates, I2a2a1a1a2a, Y3684; Vince Vaughn, actor, I2a2a1, M284; Eddie Izzard, I2a2a, L1229; Davy Crockett, I2, L801; John Tyler, 10th President, I2, L801; Ralph Waldo Emerson, Philosopher, I2, L801; Andrew Johnson, 17th president, I2a2a, L801; George Armstrong Custer, I2, L801; Chuck Norris, I2, L801.
Stephen King, I2a2a3a – L801 > Z170 > CTS6433 > S2364 > S2361 > Z78 > CTS8584 > Z185 > Z180 > L1198 > FT73935 > Y6060 > Y5748 > Y46018 > Y7272. Born September 21, 1947, renowned American author of horror, science fiction and fantasy. King has been awarded numerous prize awards and in 2003, the National Book Foundation awarded him the Medal for Distinguished Contribution to American Letters – refer Chapter XXXIV Dan: The Invisible Tribe.
House of Hohenzollern, I2a2a2a, P78 > Y7219. ‘The Hohenzollern originated from Swabia in the 11th century, became Counts of Hohenzollern in 1204, then Margraves of Brandenburg in 1411, Dukes of Prussia from 1525, Kings of Prussia from 1701, and eventually German Emperors from 1871 to 1918 under Wilhelm I and Wilhelm II.’

Ted Danson, I2a2b, L38.
Elvis Presley, b. 1935, d. 1977; I2c1a2a1a1a, F2044 – ‘Elvis’s paternal grandfather was born out of wedlock’ and received his mother’s surname. ‘His Y-DNA test showed numerous exact matches’ with the last name of Wallace in Scotland, who belong ‘overwhelmingly to the rare’ Haplogroup derived from I2c1.

An important point to understand is that labelling a paternal Y-DNA Haplogroup as purely one ethnic group can be limiting. Even so, Haplogroups can certainly be largely indicative of a specific ancestor group. Though Haplogroup I1 (M253) is characteristic of Scandinavians and Germanic peoples, in reality its origin may lay with neither. The Haplogroup tree reveals Haplogroup I as a rather old Haplogroup for European descended peoples.
Yet Haplogroup I is a bit mysterious, for which Europeans is it the paternal ancestor? Like G2a, it is a diminished Haplogroup which has been superseded by its descendant Haplogroup lines, in this case R1a and R1b. Haplogroup I1 with the U106 sub-clade of R1b and Z284 of R1a are all strongly associated with migrations of Germanic tribes from Scandinavia and northern Germany. Haplogroup I1 was close to non-existent outside of these regions. Like R1a, I1 (and I2) is a result of admixture, as it is R1b which is the defining marker Haplogroup for northwestern European men descended from Abraham.
Like R1a and R1b, Haplogroup I has split so that I2a1 is very much associated with the Balkans and southeastern Europe. Whereas (formerly) I2a2 (I2a1b1) with I1 are each reflective of western and northwestern Europe respectively. Therefore, just as an ancient ancestor carried R1 which split into R1a and R1b; the same has occurred for I1 and I2. But (as mentioned), the difference with I1 compared to say R1b in western Europeans is that the definitive defining marker Haplogroup for Scandinavians and Germans is for example R1b and not I1. A similar comparison is in eastern Europe where the defining marker Haplogroup for Slavic speaking peoples is R1a. Yet within these nations there can be quite high percentages of I2a1 carrying males.
R1b and R1a as the dominant Haplogroups in Europe overall, reveal a common paternal ancestor for R1b males, another one for R1a males and an older common ancestor for both in R1. Within these nations there are other males who possess an even older ancestor who carried Haplogroup I. So that in Sweden for instance where 21.5% of men are R1b, these are the true Swede for the want of a better word, descended from Abraham and his second wife, Keturah – Chapter XXVII Abraham & Keturah – Benelux & Scandinavia. It links them with all the other related R1b peoples in Scandinavia, the Benelux nations, Germany, Austria, Britain and Ireland.
Unlike Finland, where the predominant N1c1 is from admixture; it is I1 that is the original Haplogroup for Finns. Haplogroup I1 males in Sweden even though comprising a majority of some 37% of the population, are in reality while still ‘Swedish’, separate and distinct – even if only minutely because of centuries of intermixing – because of their decidedly different and older paternal lineage.
The question of whether I1 is Scandinavian or Germanic is similar to whether I2a1 is wholly Slavic like R1a or is it indicative of a different, older male lineage. Very like Sweden, Croatia has 24% of its males who possess R1a, yet the majority carry I2a1 with 37%. The true Croat male may carry the more recent Slavic R1a and those Croats with I2a1 – as with the Swedes who carry I1 – are from an earlier paternal ancestor and clearly not the same – by varying degrees, according to admixture. That said, it may be the other way around for the men of the Balkan nations descended from the former Yugoslavia, in that the R1a males are a spillover from the Slavic peoples of central eastern Europe and it is in fact the I2a1 males who are the true Croats and Bosnians for example.
So in a seeming contradiction, the Swedish (probably) and Croat (possibly) males who carry the older Haplogroup mutations I1 and I2a1 are not as ‘Scandinavian or Slavic’ as those exhibiting R1b and R1a respectively. It is not intended for this premise to offend anyone and it is hoped that the concept is received in its context and not misunderstood as any slight on any specific peoples within the nations used as examples. And so, a curious conundrum is that the I1 Swedish men and I2a1 Croatian men while geographically distant share a closer paternal (Y-DNA) ancestry than they do with kindred R1b and R1a Swedes and Croats living in Sweden and Croatia…
A brief summary of the Y-DNA Haplogroups surveyed thus far. Haplogroups A and B are associated with peoples of Black African heritage as are the later mutations of Haplogroup E, including E1a, E1b1a and E2. Haplogroup E1b1b is associated mainly with Berbers and related ‘non-Arab’ peoples in East Africa and those in southern Europe from admixture.
Haplogroup H is indicative of peoples in the southern portion of the Indian sub-Continent and Bangladesh. All these peoples descend from Noah’s son Ham.
Haplogroups C and D are associated primarily with Central Asians and East Asians, who descend from Noah’s eldest son Japheth.
Haplogroup F is an intersection Haplogroup for A, B, C , D and E which preceded it and those which derive from F: G, H, I and J. Haplogroup G is the first ostensibly European Haplogroup followed by the later mutations from Haplogroup I of I1 and I2. These are indicative of Shem’s descendants, the youngest son of Noah.
Put another way, Haplogroups A, B, E1b1a and E1b1b are indicative of the offspring of Canaan; while Haplogroup H of the sons of Cush. Haplogroup C is located the most frequently amongst Madai today and Haplogroup D in Tarshish, the second son of Javan. Haplogroup G is more difficult to isolate beyond Shem, whereas Haplogroup I is indicative amongst descendants of Shem’s third born son, Arphaxad.
Most of these Haplogroups, whether ancient or old have a lower frequency in the world with less mutations, which include A, B, C, D, F and G. Haplogroup H though old, is found in high concentrations, while contrastingly intermediate Haplogroup I is less concentrated with numerous sub-clades. It is Haplogroup E which stands out, as a widespread Haplogroup; one with high concentrations; and numerous mutations and sub-clades.
The final Haplogroup derived indirectly from Group F is the intermediate Haplogroup J (M304). It is not part of the M89 mutation which bonds Haplogroups F, G and H. It derives originally from the Haplogroup IJK, L15 and L16 mutations and then (arguably) IJ (M429). Thus J split from IJ and IJ and K derive from IJK. It is only at this point that IJK joins with Haplogroup G (M201) and H (L901) as immediate descendants of Haplogroup F (M89).
Haplogroup J has two main sub-groups, J1–M267 and J2–M172, believed to have arisen 10,000 years ago in Armenia and the Zagros mountains respectively. Yet chromosomes F-M89* and IJ-M429* ‘were reported to have been observed in the Iranian plateau (Grugni et al. 2012).’
Haplogroup J has also been detected in two ancient Egyptian mummies ‘excavated at the Abusir el-Meleq archaeological site in Middle Egypt, which date from a period between the late New Kingdom and the Roman Era.’ This is significant, as the sons of Mizra though not the only Egyptian dynasties or Pharaohs through history, have been the predominant rulers in later dynasties. And, we will find that Haplogroup J mirrors the demographic of E1b1b dispersal amongst Berbers and related peoples in southern Europe.
Haplogroup J-M304 is found in its greatest concentration in the Arabian Peninsula in contrast with E1b1b in North Africa. ‘Outside of this region, haplogroup J-M304 has a significant presence in other parts of the Middle East as well as in North Africa, the Horn of Africa, and Caucasus. It also has a moderate occurrence in Southern Europe, especially in central and southern Italy, Malta, Greece and Albania’ as well as in Turkey. A sub-clade of J, M140 is found in Anatolia, Greece and southern Italy, while M172 is also found in Central Asia and South Asia. Haplogroup J* (J-M304*) is rare outside the island of Socotra in Yemen.
The principal sub-clades for Haplogroup J:
J-M304 12f2a, 12f2.1, M304, P209, L60, L134
- J1 M267, L255, L321, L765, L814, L827, L1030
- M62
- M365.1
- L136, L572, L620
- M390
- P56
- P58, L815, L828
- L256
- Z1828, Z1829, Z1832, Z1833, Z1834, Z1836, Z1839, Z1840, Z1841, Z1843, Z1844
- Z1842
- L972
- J2 M172, L228
- M410, L152, L212, L505, L532, L559
- M289
- L26, L27, L927
- L581
- M12, M102, M221, M314, L282
- M205
- M241
- M410, L152, L212, L505, L532, L559
Haplogroup J1 (M267) is found in ‘Semitid/Bedouinid Arabids’ and is associated with Semitic languages speaking people in the Middle East, Ethiopia and North Africa, as well as in Mediterranean Europe though in smaller frequencies like Haplogroup T. J1 is also found in Dagestan, Iran, Pakistan and India. Haplogroup J1 highest concentrations include: the Marsh Arabs of southern Iraq, 81%; Yemen, up to 76%; Saudi Arabia, 64%; Qatar 58%; Arab Bedouins, 62%; Ashkenazi Jews, 20%; Iraq, 28%; and Egypt, 20%.
‘To some extent, the frequency of Haplogroup J-M267 collapses at the borders of Arabic/Semitic-speaking territories with mainly non-Arabic/Semitic speaking territories, such as Turkey [Elam] (9%), [and] Iran [Lud] (5%)…’ J1 ‘is also highly frequent among the… [Jewish] Kohanim [Cohen] line (46%) (Hammer 2009).’ This lands a huge blow for the contention that the Cohen line is descended from Jacob’s son, Levi – refer Chapter XXIX Esau: The Thirteenth Tribe; and Chapter XXXI Reuben, Simeon, Levi & Gad – the Celtic Tribes.
Wherever J1 and T1 are present in high frequency, mtDNA haplogroups HV, N1 and U3 are also found. To a lesser extent, Haplogroups J, K and T are also exhibited. Understandably, J-M267 as a non-European lineage is uncommon in Northern and Central Europe. Through integration and intermarriage J1 has pocket level frequencies of 5% to 10% among various populations in southern Europe.
The original or true lineage of the historic Arab people – purportedly from Jordan or Saudi Arabia – is deemed as J1-FGC12, aka S21237. The view is that this sub-clade began to spread in the Arabian Peninsula about 3,000 years ago and appeared to experience a tremendous expansion during the past 1,300 years. These ‘Arabic’ J1-FGC12 lineages ‘are found throughout the Arabic-speaking world, but they only represent a small minority of lineages in any region but the Arabian peninsula.’
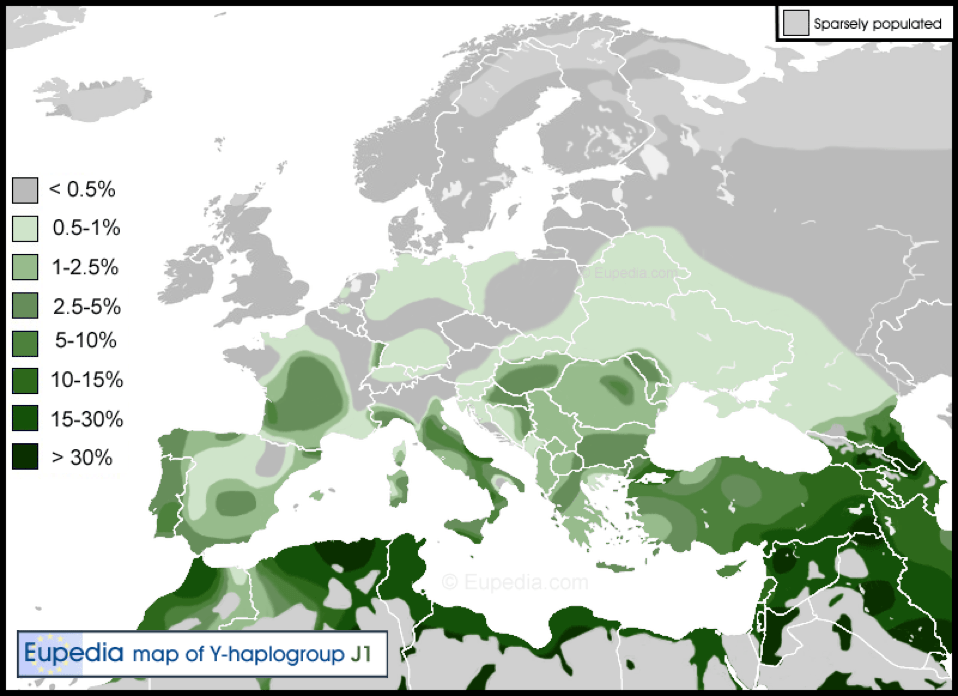
Other sub-clades of J1 ‘cannot be considered to be the paternal descendants of [the] first speakers of Arabic. These other J1 lineages were Arabicized alongside other haplogroups [such as E1b1b and] J2… during the Islamic expansion from the 7th century onward. More importantly, J1-FGC12 is not the only haplogroup that spread with the Arabic expansion linked to the diffusion of Islam. Nowadays only 40% of Saudis and 30% of Jordanians belong to J1 (most but not all to FGC12). E1b1b-M34** [through admixture] is another important Arabic lineage… found in 25% of Jordanians and 10% of Saudis.’
J1 (L255, L321, M267)
- J1* clusters are found in Eastern Anatolia and parts of the Caucasus.
- J1a (M62) Found at very low frequency in Britain.
- J1b (M365.1) Found at low frequency in Eastern Anatolia, Iran and parts of Europe.
- J1c (L136)
- J1c* Found at low frequency in Europe.
- J1c1 (M390)
- J1c2 (P56) Found sporadically in Anatolia, East Africa, the Arabian Peninsula and Europe.
- J1c3
- J1c3* Found at low frequency in the Levant and the Arabian Peninsula.
- J1c3a (M367.1, M368.1) Previously known as J1e1.
- J1c3b (M369) Previously known as J1e2.
- J1c3c (L92, L93) Found at low frequency in South Arabia.
- J1c3d (L147.1) Accounts for the majority of J1, the predominant Haplogroup in the Arabian peninsula.
- J1c3d* Accounts for the majority of J1 in Yemen, Cohen Jews (both Rabbinical and Karaitic) but missing from Quraysh including Sharif of Makkah of Banu Hashem clan.
- J1c3d1 (L174.1)
- J1c3d2 (L222.2) Accounts for the majority of J1c3d in Saudi Arabia. An important element of J1c3d in North Africa.
- J1c3d2*
- J1c3d2a (L65.2/S159.2)
- J1c3d2*
Online Encyclopaedia – emphasis & bold mine:
‘Like J1-P58, E-M34**… is… shared with their Semitic cousins, the Jews. Haplogroup E1b1b is considered the prime candidate for the origin and dispersal of Afro-Asiatic languages across northern and eastern Africa and south-west Asia. The Semitic languages appear to have originated within a subclade of the M34 branch of E1b1b. One specific deeper subclade is surely associated with the development of Arabic language and with J1-FGC12, but it hasn’t been identified yet. Note that E-M34 itself is many thousands of years old and is also found in non-Semitic countries, including Turkey, Greece, Italy, France and Spain.’

‘The two most common Jewish subclades of J1 downstream of P58 are Z18297 and ZS227. The latter includes the Cohanim haplotype. Most of the other branches under P58 could be described as Semitic, although only FGC12 seems to be genuinely linked to the medieval Arabic expansion from Saudi Arabia. J1-P58 (J1a2b on the ISOGG tree, formerly known as J1e, then as J1c3) is by far the most widespread subclade of J1. It is a typically Semitic haplogroup, making up most of the population of the Arabian peninsula, where it accounts for approximately 40% to 75% of male lineages.’
Famous male J1 individuals include: Clan Graham, J1a-P58; Dustin Hoffman, J1-Z18271 downstream of ZS227; Noah Webster Jr, American Lexicographer, J1-BY161126 downstream of L858; Alan Dershowitz, American lawyer and author.
Haplogroup J2 (M172) follows a seemingly slightly different ethnic and geographic pattern from J1 (M267). Though that said, the closeness of the two lineages supports the contention that the ‘non-Arabic’ J2 is related to the defining marker Arab Haplogroup, J1.
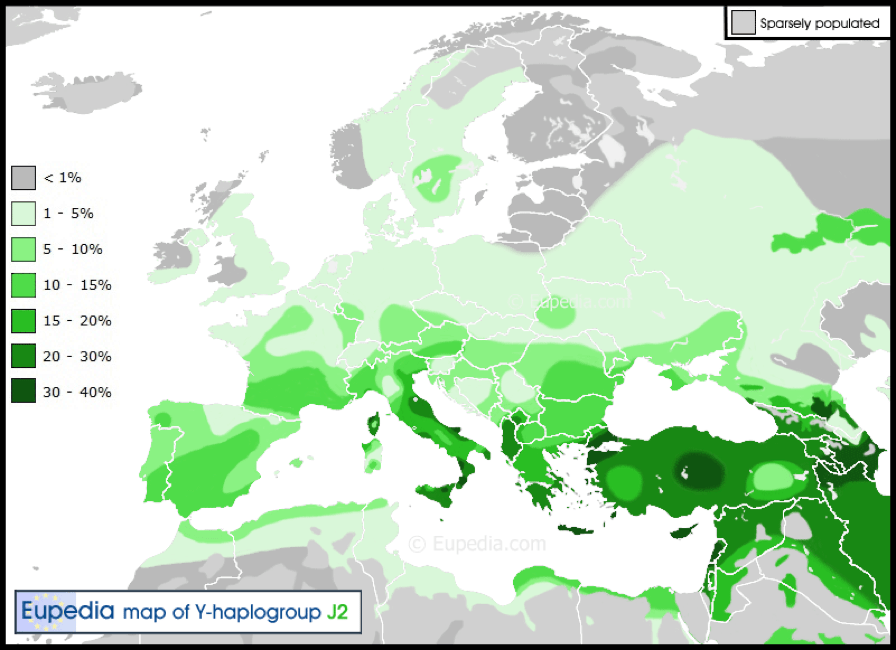
Haplogroup J2 is found primarily in ‘Syrid/Nahrainid Arabids’ located in North Africa, West Asia, Central Asia, Italy, Greece, the Balkans as well as Turkey, Iran, the Caucasus and South Asia.
J2 M172 PF4908, L228/PF4895/S321
- J2a M410, L152, L212/PF4988, L505, L532, L559/PF4986
- J2a1 DYS413≤18, L26/PF5110/S57, F4326/L27/PF5111/S396
- J2a1a M47, M322
- J2a1b M67/PF5137/S51
- J2a1c M68
- J2a1d M319
- J2a1e M339
- J2a1f M419
- J2a1g P81/PF4275
- J2a1h L24/S286, L207.1
- J2a1i L88.2, L198
- J2a2 L581/PF5026/S398
- J2a2a P279/PF5065
- J2a1 DYS413≤18, L26/PF5110/S57, F4326/L27/PF5111/S396
- J2b M12, M102, M221, M314, L282
- J2b1 M205
- J2b2 M241
- J2b2a1 L283
Encyclopaedia: ‘The world’s highest frequency of J2 is found among the Ingush ([87.4%] of the male lineages) and Chechen [55.2%] people in the Northeast Caucasus. Both belong to the Nakh ethnic group, who have inhabited that territory since at least 3000 BCE. Their language is distantly related to Dagestanian languages, but not to any other linguistic group. However, Dagestani peoples (Dargins, Lezgins, Avars) belong predominantly to haplogroup J1 (84% among the Dargins) and almost completely lack J2 lineages.
Other high incidence of haplogroup J2 are found in many other Caucasian populations, including the Azeri (30%), the Georgians (27%), the Kumyks (25%), and the Armenians (22%). Outside the Caucasus, the highest frequencies of J2 are observed in Cyprus (37%), Crete (34%), northern Iraq (28%), Lebanon (26%), Turkey (24%…), Greece (23%), Central Italy (23%), Sicily (23%), South Italy (21.5%), and Albania (19.5%), as well as among [Sephardic Jews from 15% to 29% and Ashkenazi Jewish men with 15% to 23%].
Ancient J-M410 [J2a], specifically subclade J-Y12379*, has been found, in a mesolithic context, in a tooth from the Kotias Klde Cave in western Georgia dating 9.529-9.895 cal. BP. In Italy, J-M172 is found with regional frequencies ranging between 9% and 36%… In Greece, it is found with regional frequencies ranging between 10% and 48%. Approximately 24% of Turkish men are J-M172 according to a recent study… with regional frequencies ranging between 13% and 40%… Combined with J-M267, up to half of the Turkish [male] population belongs to Haplogroup J-P209.’
It is important to remember that it is Haplogroup R1a which is the defining marker Haplogroup for Greeks and R1b for Turks and Italians. The J1 and J2 lineages are older lines of lineal descent from originally Ham and not to be equated with the R1a and R1b lineages inherited through Shem’s five sons.
‘It was reported in an early study which tested only four STR markers… that a small sample of Italian Cohens belonged to Network 1.2, an early designation for the overall clade now known as J-L26, defined by the deletion at DYS413. However, a large number of all Jewish Cohens in the world belong to haplogroup J-M267…’
‘Haplogroup J2 has been present in South Asia mostly as J2a-M410 and J2b-M102… J2-M172 was found to be significantly higher among Dravidian castes at 19% than among [northern Indian] castes at 11%. J2-M172 and J-M410 [J2a] is found [at] 21% among Dravidian middle castes, followed by upper castes, 18.6%, and lower castes 14%.
Within the Indian subcontinent, J2a peaks at frequencies of 15-25% around the Indo-Pakistani border, from Punjab to Gujarat and Sindh. In Pakistan, the highest frequencies of J2-M172 were observed among the Parsis at 38.89%… J2-M172 is found at an overall frequency of 16.1% in the people of Sri Lanka…’

Famous J2a individuals include: Clan Montgomery, J2a1-L26; Vincent van Gogh, J2a1-L26; the Rothschild Family, J2a1-Y23457 under M67, Z467 and Y15238; John Curtin, 14th Prime Minister of Australia, J2a1-F3133; Burt Bacharach, J2a1-L556/L560; Bernie Sanders, J2a; Adam Sandler, J2a1-Z30390 downstream of M67 and L210; Ben Affleck, J2a1d; Stephen Langton, Archbishop of Canterbury and author of the Magna Carta, J2a1-M319.
The Younger brothers sub-clade is J2a1h2a1-FGC24630: ‘Cole, Jim, John, and Bob Younger were notable members of the 19th-century James-Younger gang of American outlaws, which also included Jesse James. Their deep clade is downstream of L25 > L70 > Z2177 > PH185.’
Famous J2b individuals include: John Stamos, actor and singer, J2b2a-Z631; John Field, Astronomer in the Court of Elizabeth I, J2b2a-Z8429.
Haplogroup K [M9, (P128, P131, P132)] is the third intersection Y-DNA Haplogroup following CT and F. Haplogroup K derives from the Haplogroup IJK L15 and L16 mutations. Haplogroup K in turn is the parent of the groups with mutation M9. They include Haplogroup LT or K1 (L298/P326), from which L and T derive; K2 (M526) ancestor of Haplogroup NO or K2a2 (M214), from which N and O descend; Haplogroup S (B254) and Haplogroup M (M256) from K2b1; with finally, Haplogroup P or K2b2, from which Haplogroups Q (M242) and R (M207) descend.
While Haplogroup K is the ancestral parent Haplogroup of groups L to R, K also includes minor sub-Haplogroups, which are present at low frequencies in dispersed geographic regions all around the world. Haplogroup K is complex with mutational splits which include primarily descendants from Japheth (K, N, O, Q) as well as Ham (L, M, R2, S, T) and Shem (R1). Haplogroup K-M9 is spread all throughout ‘Eurasia, Oceania and Native Americans’, and found on every continent except Antartica.

The main clades of K* [LT (K1), K2a, K2b], K2c, K2d and K2e are mainly found in ‘Melanesia, Aboriginal Australians, India’ – all descended from Cush – ‘Polynesia and Island South East Asia’ – each descended from Javan. Basal K* is exceptionally rare with confirmed examples of K-M9* most common amongst a few populations in Archipelago South East Asia and Melanesia. ‘The only living males reported to carry the basal Haplogroup K2* [M256] are indigenous Australians. Major studies published in 2014 and 2015 suggest that up to 27% of Aboriginal Australian males carry K2*, while others carry a subclade of K2.’
Preceding K2 was K1, also known as LT (L298); yet it ‘has never been found in basal form (LT*).’ Sub-clades ‘are widely distributed at low concentrations.’ Haplogroup L-M20 (K1a) [M22 (L1), M317 (L1b), M349 (L1b1), M27 (L1a1), M357 (L1a2), L595 (L2)] ‘is found at its highest frequency in [southern] India, [19%], Pakistan [13%] and among the Baloch of Afghanistan [28%].’
Whereas Haplogroup T-M184 (K1b) [T1-L260, T1a-M70, T1a1-L162, T1a2-L131, T1a3 (T2)] ‘is most common among: Fulanis, Toubou, Taureg, Somalis, [the Horn of Africa amongst Cushitic-speaking peoples] Egyptians, Omanis, some [inhabitants in the] Middle East, Sephardi Jews, the Aegean Islands and among Kurru, Bauris and Lodha in India.’ Haplogroup T, while geographically widespread, is relatively rare. Maternal lineages associated with T include: HV, N1a and U3 (all Arab). The third United States President, Thomas Jefferson belonged to Haplogroup T1a1a-L208.
Haplogroup K2a (M2308) derives from K2. ‘K2a* – found only in the remains of Ust’-Ishim man… found in Omsk Oblast, Russia… [and] were initially classified, erroneously, as K2*…’ K-M2313* has only been found in one Telugu male and in one ethnic Malay. From K-M2313, Haplogroup NO (M214) or K2a2 mutated.
Branching off from K2a is K2b (P331) and then K2b1 which is the parent of Haplogroups S (B254) and M (P256). Also descending from K2b is K2b2 or Haplogroup P, which is the fourth and final intersection Haplogroup. Other sub-clades branching off from K2 include: K2c (P261), a minor lineage found in Bali, Indonesia; K2d (P402), also a minor lineage, found in Java, Indonesia; and K2e (M147), a rare lineage located in South Asia.
Haplogroup N (M231) and N1c (L729) [including N1c2 (L666)] has a wide geographic distribution amongst populations throughout northern Eurasia, including China, North and South Korea, Japan, Mongolia and particularly Uralic speakers of northern Siberia, as well as Central Asia.
‘Haplogroup N1c[1] is found chiefly in north-eastern Europe, particularly in Finland (61%), Lapland (53%), Estonia (34%), Latvia (38%), Lithuania (42%) and northern Russia (30%), and to a lower extent also in central Russia (15%), Belarus (10%), eastern Ukraine (9%), Sweden (7%), Poland (4%) and Turkey (4%). N1c is also prominent among the Uralic speaking ethnicities of the Volga-Ural region, including the Udmurts (67%), Komi (51%), Mari (50%) and Mordvins (20%), but also among their Turkic neighbours like the Chuvashs (28%), Volga Tatars (21%) and Bashkirs (17%), as well as the Nogais (9%) of southern Russia.’
Haplogroup N1c1 is strongly associated with Uralic peoples through admixture, which is divided in the following families.
- Samoyedic (Nganasans, Enets, Nenets and Selkups)
- Finno-Ugric
- Finno-Permic
- Baltic Finnic (Finnish, Karelian, Estonia, etc.)
- Permic (Komi, Udmurt)
- Saamic (Saami)
- Volgaic (Mari, Mordvin)
- Ugric
- Hungarian
- Ob-Ugric (Khanty, Mansi)
- Finno-Permic
The most frequent sub-clade of L729 is N1c1 (M46). ‘It probably arose in a Northeast Asian population, because the oldest ancient samples comply with this genetic profile. [Haplogroup] N has experienced serial bottlenecks in Siberia and secondary expansions in eastern Europe…’ Though certain sub-clades are very common in Finland and the Baltic nations comprising Estonia, Latvia and Lithuania, the origin of Haplogroup N sits squarely as a lineage from Japheth. For instance, ‘in Siberia, haplogroup N-M46 reaches a maximum frequency of approximately 90% among the Yakuts, a [Turko-Mongol] people who live mainly in the Sakha… [a] Republic [in Russia].’
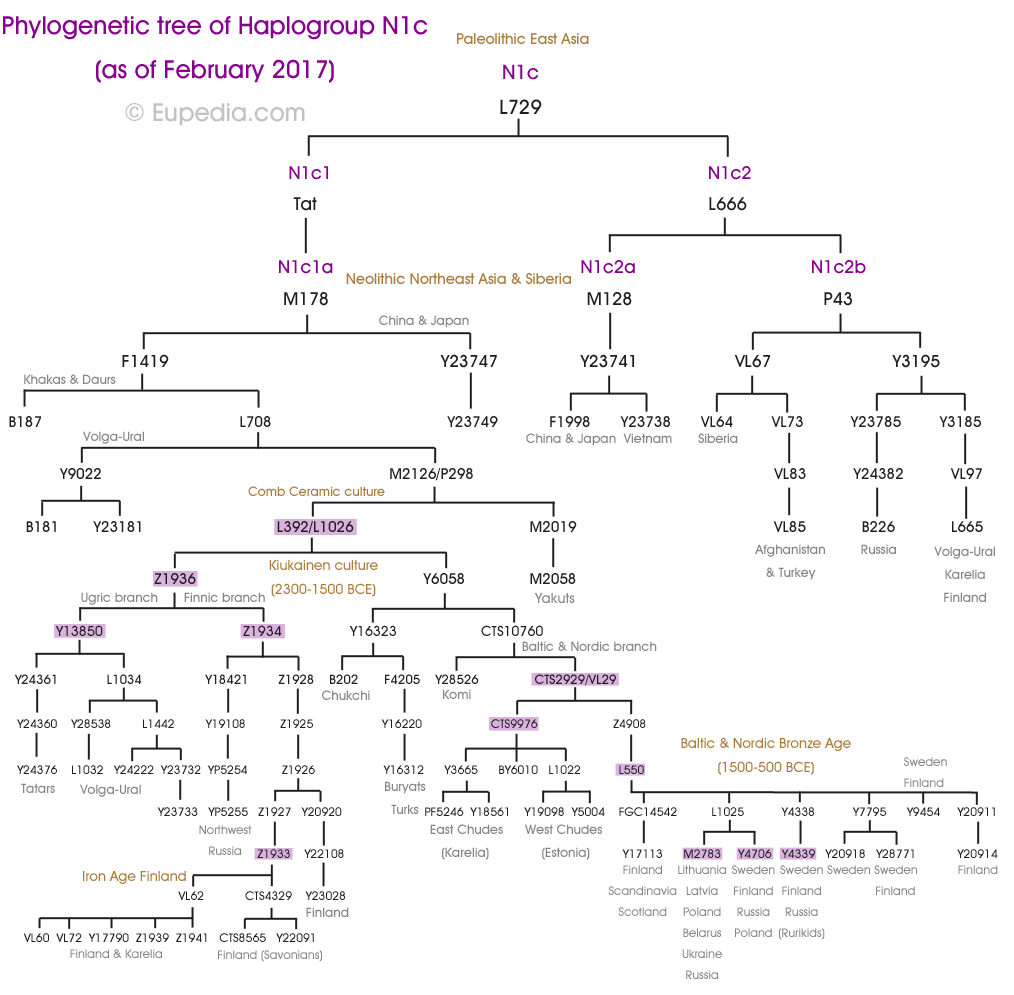
Unlike Haplogroups I and J which are associated with Shem and Ham respectively; Haplogroups N and O are both lines of descent deriving from Japheth. In Finland the two main patrilineal Haplogroups are N1c at 61.5% and I1 with 28%. The contention is that the Finnish men with N1c1a (M178) are an intertwined Japheth line, whereas those with I1 represent if not the true Finnish male, a more ancient unmixed Finn descending from Shem. Similarly, the 32% of men in Estonia with R1a perhaps represent an original Estonian line of descent compared with the 34% of men with Haplogroup N.
Paternal Haplogroup O (M175) is a major defining marker Haplogroup for the descendants of Japheth. Lineage O represents nearly 60% of chromosomes for males in East Asia and it is numerically dominant throughout East Asia, Southeast Asia and by degree in the South Pacific and Central Asia. There are some 1.4 billion Chinese, with the peoples of East Asia and South East Asia numbering approximately another 870 million. Adding the populations for these two regions together and then dividing them in half, provides an approximate figure for the male populations. It is a staggering amount of people, yet there is one other group which outnumbers O and that is Haplogroup R.

Haplogroup O descends from NO-M214 and has two main branches identified as O1 (F265), also known as F75 and O2 (M122). Haplogroup O1 divides again into the primary lineages O1a (M119) and O1b (M268, P31). One source puts forward that ‘O1-F265 should have existed as a single haplogroup parallel to O2-M122 for a duration of approximately 762 years (or anywhere from 0 to 13,170 years considering the 95% CIs and assuming that the phylogeny is correct) before breaking up into its two extant descendant haplogroups, O1-MSY2.2 and O1b-M268.’
Haplogroup O-M175 appears in 80% to 90% of most populations in both East Asia and Southeast Asia. Plus it is almost exclusive to this region of the world as a massive marker for Japheth’s sons, Magog, Tubal, Meshech, Gomer and Javan. Haplogroup O is virtually non-existent in the rest of the world except through migration and inter-marriage. ‘However, certain subclades of Haplogroup O-M175 do achieve significant frequencies among some populations of Central Asia, South Asia, and Oceania. For example, one study found it at a rate of 65.81% among the Naimans, a tribe in Kazakhstan, even though the rate among Kazakhs in general is believed to be only about 9%…’
Haplogroup O is associated with the spread of Austronesian languages. For example, Haplogroup O-M50 has even been found with O-M95(xM88) among the Malagasy people of Madagascar with a combined frequency of 34% – Chapter VII Javan: Archipelago South East Asia & Polynesia. Haplogroup O-M175 is found in 88.7% of Asian Americans; 1.6% in Hispanic Americans; 0.5% in White Americans; and 0.3% to 0.5% in African Americans.
The first of the three major sub-Haplogroups for group O is O1a (M119). It is found principally in the populations of southeastern China [Tubal], Taiwan [(Tubal), Javan-Rodan], Malaysia [Javan-Elishah], Indonesia [Javan- Kittim], the Philippines [Javan-Dodan], the Pacific Islands [Javan-Rodan] and Madagascar.
Haplogroup O1a is associated with the spread of the Austronesian languages, including Formosan and Malayo-Polynesian as well as Kra-Dai and Tai. The majority of these peoples are associated with Japheth’s fourth son, Javan and three out of four of his sons.
High frequencies of O1a have been found in populations ‘spread in an arc through southeastern China, Taiwan, the Philippines, and Indonesia. It has been found with generally lower frequency in samples from Oceania, mainland Southeast Asia, Southwest China, Northwest China, North China, Northeast China, Korea, Japan, North Asia, and Central Asia.’ Haplogroup O1a occurs in a low average frequency of about 4% among the Han populations of northern China. Whereas the peoples of southwestern China and Southeast Asia who speak Tibeto-Burman languages and in the Han population, show a higher frequency of carrying between 15% to 23% O1a.
One source ‘suggests that modern Southern Han populations may possess a non-trivial number of male ancestors who were originally affiliated with some Austronesian-related culture, or who at least shared some genetic affinity with many of the ancestors of modern Austronesian peoples’ – Chapter X China: Magog, Tubal & Meshech.
A link between Tubal, the southeastern Chinese and Javan, Archipelago South East Asia is supported by Karafet’s 2005 findings – Ezekiel 27:13. ‘This lineage is found frequently in Austronesians, southern Han Chinese, and Kra-Dai peoples… [and] is presumed to be a marker of the prehistoric Austronesian expansion, with possible origins encompassing the regions along the southeastern coast of China and neighboring Taiwan, and is found among modern populations of Maritime Southeast Asia and Oceania.’
The second of the three major sub-Haplogroups of group O is O1b (M268, P31). O1b exhibits a less uniform dispersal than O1a and is somewhat peculiar in its geographic distribution. It is associated with the spread of the Austroasiatic languages (O1b1a1a-M95), Munda and Mon-Khmer. Found amongst Tai peoples [Gomer-Minni], Hlai, Balinese, Javanese [Javan-Kittim], Malagasy, Manchus, Ryukyuans, Japanese [Javan-Tarshish] and Koreans [Gomer-Togarmah]. O1b concentration aligns with the descendants of Gomer’s sons and two of Javan’s four sons – and not so much with Magog, Tubal and Meshech of China.
Encyclopaedia:
‘… Haplogroup O-P31 is generally found with high frequency only among certain populations, such as the Austroasiatic peoples of India, Bangladesh and Southeast Asia, the Nicobarese of the Nicobar Islands in the Indian Ocean, Koreans, and Japanese. Besides its widespread and patchy distribution, Haplogroup O1b-P31 is also notable for the fact that it can be divided into three major subclades that show almost completely disjunct distribution. One of these subclades, O1B1-K18 can be mainly divided into two subclades O1b1a1-PK4 (formerly O2a) and O1b1a2… (formerly O2*(xM95,M176)).
O1b1a1-PK4 is found among some (mostly tribal) populations of South and Southeast Asia, as well as among the Japanese… Javanese, Sundanese, and Balinese of Indonesia and some Zhurong related Chinese.’ The link between the Japanese and peoples of Malaysia and Indonesia is addressed in Chapter VII Javan: Archipelago South East Asia & Polynesia; and Chapter IX Tarshish & Japan.
‘O1b1a2… is relatively rare and mainly distributed in East Asia, especially in some Yue, Baiyue related Chinese. Another subclade, Haplogroup O1b2-M176 (formerly O2b), is found almost exclusively among the Japanese, some Buyeo Koreans and Jin Manchurians. A broad survey of Y-chromosome variation among populations of central Eurasia found haplogroup O-M175(xM119, M95, M122) in 31% (14/45) of a sample of Koreans… However, nearly all of the purported Korean O-M175(xM119, M95, M122) Y-chromosomes may belong to Haplogroup O-M176 and later studies do not support the finding of O-M175* among similar population samples… The reported examples of O-M175(xM119, M95, M122) Y-chromosomes that have been found among these populations might therefore belong to Haplogroup O-M268*(xM95, M176) or Haplogroup O-M176 (O1b2).’
The third of the three major sub-Haplogroups for group O is O2 (M122). Haplogroup M122 is primarily associated with Chinese people (Magog, Meshech, [Tubal]), yet it forms a substantial component of the Y-chromosome diversity in many modern populations of the East Asian region. Haplogroup O2 is associated with the spread of Sinitic and Tibetan-Burman languages (O2a2b1-M134), as well as Hmong and Mien languages – O2a2a1a2 (M7). ‘Haplogroup O-M122 comprises about 50% or more of the total Y-chromosome variation among the populations of each of these language families.
The Sinitic and Tibeto-Burman language families are generally believed to be derived from a common Sino-Tibetan protolanguage, and most linguists place the homeland of the Sino-Tibetan language family somewhere in northern China. The Hmong–Mien languages and cultures, for various archaeological and ethnohistorical reasons, are also generally believed to have derived from a source somewhere north of their current distribution, perhaps in northern or central China.’

Haplogroup O2 formerly O3, ranges across East Asia and South East Asia where it dominates the paternal lineages with extremely high frequencies.
Online Encyclopaedia: ‘Researchers believe that O-M122 first appeared in Southeast Asia… In a systematic sampling and genetic screening of an East Asian–specific Y-chromosome haplogroup (O-M122) in 2,332 individuals from diverse East Asian populations, results indicate that the O-M122 lineage is dominant in East Asian populations, with an average frequency of 44.3%. Microsatellite data show that the O-M122 haplotypes are more diverse in Southeast Asia than those in northern East Asia. This suggests a southern origin of the O-M122 mutation to be likely [rather northern]. However, the prehistoric peopling of East Asia by modern humans remains controversial with respect to early population migrations and the place of the O-M122 lineage in these migrations is ambivalent.
Haplogroup O-M122 is found in approximately 53.31% of all modern Chinese males… about 40% of Manchu, Chinese Mongolian, Korean, and Vietnamese males, about 33.3% to 62%… of Filipino males, about 10.5% to 55.6% of Malaysian males… about 25%… of Indonesian males, and about 16% to 20% of Japanese males’, 25% to 32.5% of Polynesian males, 18% to 27.4% of Micronesian males and 5% of Melanesians [Cush].
‘Haplogroup O-M122* Y-chromosomes, which are not defined by any identified downstream markers, are actually more common among certain non-Han Chinese populations than among Han Chinese ones, and the presence of these O-M122* Y-chromosomes among various populations of Central Asia, East Asia, and Oceania is more likely to reflect a very ancient shared ancestry of these populations rather than the result of any historical events. It remains to be seen whether Haplogroup O-M122* Y-chromosomes can be parsed into distinct subclades that display significant geographical or ethnic correlations.’
The third paragraph supports the premise that China is principally composed of three primary paternal lines from Magog, Tubal and Meshech, identified by perhaps the variant O2 Haplogroups for non-Han Chinese, northern Han Chinese and southern Han Chinese.
A comparison of select countries who possess Haplogroups K, O1a, O1b and O2 with percentage levels.
Japan: K [2%] – O1a [2%] – O1b [33%] – O2a [19%]
N & S Korea: K [4%] – O1a [3%] – O1b [33%] – O2a [42%]
China: K [1%] – O1a [13%] – O1b [12%] – O2a [56%]
Taiwan: O1a [2%] – O1b – [9%] – O2a – [58%]
Vietnam: O1a [6%] – O1b [33%] – O2a – [40%]
Malaysia: K [8%] – O1a [8%] – O1b [32%] – O2a [28%]
Philippines: K [20%] – O1a [28%] – O1b [3%] – O2a [39%]
Most nations do not always exhibit a preceding intersection Haplogroup, yet in East Asia the Haplogroup O males invariably possess percentage levels of their parent Haplogroup K. In Taiwan and Vietnam it is absent and for Japan, the Koreas and China it is low. In Malaysia the levels are higher, while the Filipinos stand out with 20% of males carrying Haplogroup K. These two nations, both descend from Javan and have a larger number of men with an older Haplogroup than O.
The Philippines is dominant in Haplogroup O2a (M324) yet with a high proportion of O1a too. Apart from Japan and Malaysia, the other nations surveyed are all dominant in O2; particularly China and Taiwan. Malaysia like Japan has a higher frequency of O1b, yet in Malaysia it marginally beats O2. Though in Japan unlike the others and only mirrored in Tibet, it has a higher level still of the ancient Haplogroup D1a2 with 39%. All the nations selected have a similar high level of O1b, except China, Taiwan and the Philippines. In contrast, all have lower levels of O1a, except China and especially the Philippines.
In Japan, there appears to be two distinct peoples, as evidenced by the two paternal Haplogroups D1 and O1b. One which has fairer skin and more aquiline facial features and one with tawny skin and broader body attributes. The Koreans like the Vietnamese both descend from Gomer and are split between O2 and then O1b. Which is the defining marker Haplogroup is not clearly ascertained. Malaysia is similar, yet with O1b edging O2.
The Philippines are a bit of an anomaly with such a high ratio for Haplogroup K and a far higher percentage of O1a than all the other nations. China and Taiwan are clearly O2 driven. Haplogroup O2 is prevalent in all the nations whether it is dominant or not, ranging from 19% in Japan to 56% in China. It would seem possible that other Eastern Asian nations like Japan, have more than one distinct lineage as a common denominator. So that in the Koreas and Vietnam it is O2, then O1b. In Malaysia, O1b then O2; and in the Philippines, O2 and then O1a.
What does all this tell us? Observing the overall pattern for Haplogroup O in these eight selected nations, O1a is more prevalent in Archipelago South East Asia (Javan); O1b is more prevalent outside China (Magog, Tubal, Meshech) and then mainland Asia, diminishing in the island nations and especially heading towards the Southeast; and O2 is more prevalent on the Asian mainland (Gomer) and particularly in China.
Next on the K-M9 Haplogroup tree is Haplogroup S (B254). Unlike Haplogroups NO, N and O which derive from K-M2313, which in turn mutated from K2a (M2308), Haplogroup S derives from K2b1, which descends from K2b, P331.
Haplogroup S is only found in a specified geographic area where it is ‘numerically dominant in the highlands of Papua New Guinea: subclades of S1, such as S1a3 (P315) and S1a1a1 (P308), have also been reported at levels of up to 27% among indigenous Australians, while S1a (P405; previously K2b1a) has also been found at significant levels in other parts of Oceania. S2 (P336; previously K2b1b) has been found on Alor, Timor and Borneo… S3 (P378; previously K2b1c) [is] found among Aeta people of the Philippines.’
Encyclopaedia: ‘Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome Phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. Later, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being above all timely.’
Haplogroup S1a1b (M230, P202, P204) was ‘demoted’ in 2016 from its previous position as the basal Haplogroup S* (K2b1a4). From 2002 to 2008, Haplogroup S* was known as Haplogroup K5. S-M230 is found principally in New Guinea and at lower frequencies in Melanesia and eastern Indonesia. It is the most numerically significant sub-clade of Haplogroup S1a. A study reported Haplogroup S1a1b in ‘52% (16/31) of a sample from the Papua New Guinea Highlands; 21% (7/34) of a sample from the Moluccas; 16% (5/31) of a sample from the Papua New Guinea coast; 12.5% (2/16) of a sample of Tolai from New Britain… [and] 2% (2/89) of a sample from the West New Guinea lowlands’ and its coast.
Haplogroup S is associated with Melanesian peoples in South East Asia and the Pacific. The Melanesians are related to the peoples of India, Sri Lanka and Bangladesh – refer Chapter XIII India & Pakistan: Cush & Phut. The Melanesians have also intermarried with the Polynesian peoples which is discussed in Chapter VII Javan: Archipelago South East Asia & Polynesia.
Closely related to Haplogroup S and a second branch from K2b1 is Y-DNA Haplogroup M* [P256]. Haplogroup M is also known as K2b1b and previously as K2b1d. It is the most common paternal Haplogroup in West Papua and Papua New Guinea. Is is also found among indigenous Australians and parts of Melanesia and Polynesia. Haplogroup M With Haplogroup S (B254) is the only primary sub-clades of K2b1, also known as MS.
Haplogroup P256 is found at low frequencies in New Guinea and Flores. Haplogroup M is divided into three main sub-clades. M1 (M4) which is found frequently in New Guinea and Melanesia and less frequently in Indonesia, Micronesia and Polynesia. A study by Kayser in 2003 found frequencies of 77.5% in West Papua lowlands and coasts, 74.5% in the highlands; 29% in Papua New Guinea coasts and 35.5% in the highlands. An M1 sub-clade M1b1 (M104) is found in New Guinea, Fiji, Tonga and Samoa and M2 (M353) is found in Fiji, as is M3 (P117).

Though Haplogroup P (P295) also known as K2b2 is a second branch from K2b (MPS; P331), it is also an intersection Haplogroup and the fourth and final one of the three which preceded it: CT, F and K. Haplogroup P has two primary branches: P1 (M45) and P2 (B253). There is considerable speculation regarding the geographic genesis of Haplogroup P.
Encyclopaedia: ‘Karafet et al. 2015 suggests an origin and dispersal of haplogroup P from either South Asia or Southeast Asia as part of the early human dispersal, based on the distribution of subclades now classified as P2, and more ancient clades such as K1 and K2. Hallast, Agdzhoyan, et al. concluded that the ancestral Eurasian haplogroups C, D, and F, either expanded from the Middle East or from Southeast Asia. Based on the modern distribution of basal lineages, the authors propose Southeast Asia as [the] place of dispersal for all Eurasian lineages, before the split between West-Eurasian and East-Eurasian (including Oceanian) populations. According to a study by geneticist Spencer Wells, haplogroup K, from which haplogroup P [descends], originated in the Middle East or Central Asia. It is likely that haplogroup P diverged somewhere in South Asia into P1, which expanded into Siberia and Northern Eurasia, and into P2, which expanded into Oceania and Southeast Asia.’
As discussed in Chapter I Noah Antecessor Nulla, the landing of the Ark – during the post-flood descent of sea water – was in the Himalayan Mountain range towards modern day Kashmir. Significant re-settlement after the flood began in the Indus Valley, with movement of Noah’s sons and grandson’s descendants heading west to Mesopotamia, Anatolia and Egypt. Therefore hypotheses of Haplogroup origination in southeast Asia are incorrect. A chronological order of South Asia to the Middle East and then Central Asia and beyond is accurate.
Haplogroup P has been detected at low frequencies in the Caucasus and India; with P* also found in 28% of the Aeta men of the Philippines and 10% in Timor. Haplogroup P1 (M45, PF5962) with basal P1*, known as K2b2a is located in Central Asia and Siberia. P1 is found between 22.2% and 35.4% in Tuvan men as well in the Andamanese peoples of India. The only primary sub-clades of P1 are Haplogroup Q (M242) and Haplogroup R (M207). These Haplogroups comprise most of the male lineages among Native Americans, Latino-Hispano America, Europeans and parts of Central Asia and South Asia. ‘It is possible that many cases of haplogroup P1 reported in Central Asia, South Asia and/or West Asia are members of rare or less-researched subclades of haplogroups R2 and Q, rather than P1* per se.’ Haplogroup P2 (B253) is extremely rare and has only been found in the Aeta of Luzon in the Philippines.
Haplogroup Q (M242) is found in varying levels throughout Asia – particularly in Central Asia and Siberia – Europe and the Middle East. While in the Americas Q1a3a (M3) is the dominant Y-DNA Haplogroup amongst the Amerindian – Chapter II Tiras the Amerindian. M242 has one primary sub-clade, Haplogroup Q1 (L232/S432).
2008 ISOGG tree
- Q (M242)
- Q* India, Pakistan, Afghanistan
- Q1 (P36.2) Iran
- Q1*
- Q1a (MEH2)
- Q1a*
- Q1a1 (M120, M265/N14) Found with low frequency among Bhutanese, Dungans, Han Chinese, Japanese, Koreans, Vietnamese, Mongolians, Naxi and Tibetans
- Q1a2 (M25, M143) Found at low to moderate frequency among some populations of Southwest Asia, Central Asia and Siberia
- Q1a3 (M346)
- Q1a3* Found at low frequency in Pakistan, India and Tibet
- Q1a3a (M3) Typical of indigenous peoples of the Americas
- Q1a3a*
- Q1a3a1 (M19) Found among some indigenous peoples of South America, such as the Ticuna and the Wayuu
- Q1a3a2 (M194) South America
- Q1a3a3 (M199, P106, P292) South America
- Q1a4 (P48)
- Q1a5 (P89)
- Q1a6 (M323) Found in a significant minority of Yemeni Jews
- Q1b (M378) Found at low frequency among samples of Hazara and Sindhis. Widely distributed in Europe, South Asia and East Asia. Includes Mongols, Japanese and Uyghurs of north western China. Sub-branches of sub-clade L245, Y2200 and YP1035 belong to Ashkenazi Jews. While Sephardic Jews belong to other sub-clades of L245, BZ3900, YP745 and YP1237. Q1b has also been found in Panama, Central America and the Andean region in South America.
Encyclopaedia: ‘In Y chromosome phylogenetics, subclades are the branches of a haplogroup. These subclades are also defined by single-nucleotide polymorphisms (SNPs) or unique-event polymorphisms (UEPs). Haplogroup Q-M242, according to the most recent available phylogenetics has between 15 and 21 subclades. The scientific understanding of these subclades has changed rapidly. Many key SNPs and corresponding subclades were unknown to researchers at the time of publication [and] are excluded from even recent research. This makes understanding the meaning of individual migration paths challenging.’
While Haplogroup P is an intersection Haplogroup, those men who carry P1 and P2 visibly descend from Japheth and so it applies with Haplogroup Q, like Haplogroup N before it, that Haplogroup Q is found in populations other than from Japheth, yet remains a defining marker Haplogroup for Japheth’s descendants from his seventh son, Tiras – Article: Seventh Son of a Seventh Son.
‘It is unclear whether the current frequency of Q-M242 lineages represents their frequency at the time of immigration [by the major founding groups to the Americas] or is the result of the shifts in a small founder population over [a long period of] time. These… groups of founders must have included men from the Q-M346 [Q1a3], Q-L54 (Q-Z780), and Q-M3 lineages. In North America, two other Q-lineages also have been found. These are Q-P89.1 (under Q-MEH2) [Q1a5] and Q-NWT0. They… instead [came] from later immigrants…’
Amongst the native Americans of North America, Q-M242 is found in Na-Dene speakers with an average rate of 68%. The highest frequencies include the Navajo with 92.3%; the Apache with 78.1%; and the North American Eskimo populations with about 80%. Q-M3 accounts for 46% of the men with Haplogroup Q in North America. ‘Q-M242 is estimated to occupy 3.1% of the whole US population in 2010.’ Whereas Haplogroup Q-M242 has been found in approximately 94% of Indigenous peoples of Mesoamerica and South America.
The frequencies of Q among the male population of Central and South American countries:
- 61% in Bolivia
- 51% in Guatemala
- 40.1% to 50% in Peru
- 37.6% in Ecuador
- 37.3% in Mexico
- 31.2% in El Salvador
- 15.3% to 21.8% in Panama
- 16.1% in Colombia
- 15.2% in Nicaragua
- 9.7% in Chile
- 5.3% to 23.4% in Argentina
- 5% in Costa Rica
- 3.95% in Brazil
In Siberian Tatars, the Ishtyako-Tokuz sub-group of the Tobol-Irtysh peoples have a frequency of Q-M242 at 38%. The highest frequencies of Q-M242 in Eurasia are observed in Kets in central Siberia with 93.8%. Various subgroups of Q-M242 are observed in Mongolia such as Q1a1, Q1a2 and Q1b, for an average frequency in male Mongols of about 4% to 5%. Most of the peoples in East Asia belong to sub-clade Q1a1 (M120) and across northern China it is found in 4.5% of the men. In southern China it decreases to about 2%. Amongst the Uyghurs it is 15.38%; in South Korea it is 1.9% of the male population; in Japan 0.3%; and between 0.3 to 1.2% for Taiwanese men.
Haplogroup Q shows lower frequencies overall in Southeast Asia: 5.4% in Indonesia; approximately 4% in Vietnam; 3.1% in the Philippines; 2.8% in Myanmar; and 2.5% in Thailand. In Central Asia, Haplogroup Q is found between 2% to 6% in Kazakh men; 5% to 6% of Tajiks; 5.5% of Uzbeks; and 6.9% of Afghans.
In Iran, Haplogroup Q averages 5.5%; in Saudi Arabia, 2.5%; in Syria, 1.1%; 2% in the Lebanese; and 2% in Turkey. In Pakistan, Haplogroup Q is found at 2.2%, while in India it is 2.38%; in Sri Lanka it is 3.3%; and in Tibet, 3.2%.
In central to eastern Europe, Haplogroup Q averages 1.7%. In northern Europe it can be higher such as in Sweden at 2.5% and in southern Europe, lower at around 0.5% to 1%. Amongst Ashkenazi Jewish men, Haplogroup Q (M378/L245) averages 5.2%; and in Sephardic Jews, it ranges between 2.3% to 5.6%. Combining the data, Q-M242 is ‘estimated to be in about 3.1% of males of the world.’
Famous Haplogroup Q individuals host a variety of people including a number of Jews, such as J Robert Oppenheimer a theoretical physicist, who played a major role as the Director of the Manhattan Project and the Atomic bomb – Article: Nuclear Nefariousness.

As well as the Oppenheim Family, ‘a German-Jewish… family [of Barons] which has been a prominent family in banking and finance… since at least the 18th century. According to Forbes magazine’s Family Dynasties, the Oppenheim Family divides control of their multibillion-dollar fortune among 46 family members.’
Apart from Haplogroup Q deriving from Haplogroup P1 (M45), Haplogroup R (M207) also mutated from P1. Haplogroup R mutated into R2 (M479) and R1 (M173) with R1 diverging into R1a (M420) and R1b (M343). Haplogroup R is both numerous and widespread, like Haplogroup O.
Encyclopaedia: ‘Only one confirmed example of basal R* has been found, in… old remains, known as MA1, found… near Lake Baikal in Siberia… While a living example of R-M207(xM17, M124) was reported in 2012, it was not tested for the SNP M478; the male concerned – among a sample of 158 ethnic Tajik males from Badakshan, Afghanistan – may therefore belong to R2. It is possible that neither of the primary branches of R-M207, namely R1 (R-M173) and R2 (R-M479) still exist in their basal, original forms, i.e. R1* and R2*.
No confirmed case, either living or dead, has been reported in scientific literature… Although in the case of R2*, relatively little research has been completed. Despite the rarity of R* and R1*, the relatively rapid expansion – geographically and numerically – of subclades from R1 in particular, has often been noted: “both R1a and R1b comprise young, star-like expansions” The wide geographical distribution of R1b, in particular, has also been noted.’
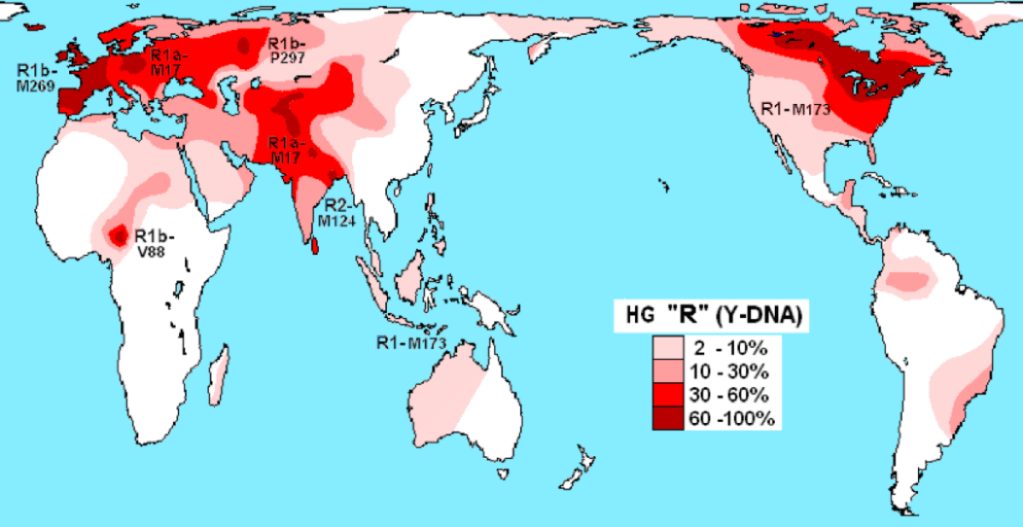
Haplogroup R2a (M124) is geographically concentrated in India, Sri Lanka and Pakistan. Its highest levels have been found in the Burusho people in northern Pakistan. Low levels of R2 are found in Iran, Anatolia, the Caucasus, Central Asia and Europe. A rare sub-clade of R-M124 is found amongst Ashkenazi Jews. Tests on R2a and R2b are still in their infancy, with further studies required. ‘The paragroup for the R-M479 lineage is found predominantly in South Asia, although deep-rooted examples have also been found among Portuguese, Spanish, Tatar (Bashkortostan, Russia), and Ossetian (Caucasus) populations…’

The colour pink represents Haplogroup R1a dispersion and red, Haplogroup R1b.
Haplogroup R1 is the most common Haplogroup in Amerindians following Haplogroup Q. Though the reasons for high levels of R-M173 among Native Americans remains a matter of controversy, as some scholars claim it is the result of ‘colonial-era migration’ from Europe; while other authorities point to the similarity of R-M173 sub-clades found in North America to those found in Siberia, supporting ‘prehistoric immigration’ from Asia. R1 is found throughout western Eurasia, yet its origins ‘cannot currently be proved.’
Haplogroup R1a (M420) is found in descendants of Shem in Eastern Europe and Scandinavia (Balto-Slavic), yet it is puzzling for it is also found in South Asia (proto-Indo-Iranian) and Central Asia, where descendants of Ham and Japheth dwell respectively. Haplogroup R1b (M343) is principally found in Western Europe and their descendants in the Americas; while sparsely represented in Asia and Africa. The marker sub-clade R1b1a1b (M269) is associated with the Italo-Celtic and Germanic peoples.
The combined R1a and R1b peoples include: India, 1.4 billion; South Asia, 370 million; Europe, 750 million; North America, 370 million; and Latino-Hispano America of 670 million, with a total of approximately 3.5 billion. Divided in half for the respective male percentage, results in Haplogroup R being far more prevalent, than the second most common Haplogroup O.
While R1a is thought to have originated during the Last Glacial Maximum, its sub-clade M417 (R1a1a1) may have diversified into Z282 (Eastern Europe) in Slavic speakers as well as Z93 (South Asia) and speakers of an Indo-Iranian language as recently as circa 5,800 years ago – Underhill, 2014. The place of origin for the sub-clade ‘plays a role in the debate about the origins of Proto-Indo-Europeans… and may also be relevant to the origins of the Indus valley civilisation.’
It is assumed to have occurred in eastern Turkey and northern Iran. Of course the location is not key, but rather with whom. In this instance the R1a mutations have been carried by at least two of the five sons of Shem, in Asshur and Arphaxad by Joktan. Similarly, R1b has been carried by descendant’s of Aram and Arphaxad through Peleg. Elam (Turkey) and Lud (Iran) each exhibit lower R1a and R1b frequencies and are not so easy to delineate.
Encyclopaedia: ‘The SNP mutation R-M420 [R1a] was discovered after R-M17 (R1a1a), which resulted in a reorganization of the lineage in particular establishing a new paragroup (designated R-M420*) for the relatively rare lineages which are not in the R-SRY10831.2 (R1a1) branch leading to R-M17.’ According to Pamjav (2012), ‘Inner and Central Asia is an overlap zone for the R1a1-Z280 [Central and Eastern Europe] and R1a1-Z93 [South Asia] lineages [which] implies that an early differentiation zone of [R1a1a]-M198 conceivably occurred somewhere within the Eurasian Steppes or the Middle East and Caucasus region as they lie between South Asia and Central and Eastern Europe. A study proposes that R1a in South Asia originally expanded from a single Central Asian source as there are at least three ‘R1a founder clades within the Subcontinent, consistent with multiple waves of arrival’ – Silva, 2017.
‘South Asian populations have the highest STR [Short Tandem repeats] diversity within R1a1a, and subsequent older TMRCA [time to most recent common ancestor] datings… From these findings some researchers have concluded that R1a1a originated in South Asia, excluding a more recent, yet minor, genetic influx from Indo-European migrants in northwestern regions such as Afghanistan, Balochistan, Punjab, and Kashmir… Sengupta also described […]:
“We found that the influence of Central Asia on the pre-existing gene pool was minor. The ages of accumulated microsatellite variation in the majority of Indian haplogroups exceed 10,000-15,000 years, which attests to the antiquity of regional differentiation. Therefore, our data do not support models that invoke a pronounced recent genetic input from Central Asia to explain the observed genetic variation in South Asia.”
‘This suggests that the origins of paternal haplogroup R1a point to the Indian subcontinent and not Central Asia. Part of the South Asian genetic ancestry derives from west Eurasian populations, and some researchers have implied that Z93 may have come to India via Iran and expanded there during the Indus Valley civilisation… [so that] the roots of Z93 lie in West Asia… [with] “Z93 and L342.2 [expanding] in a southeasterly direction from transcaucasia into South Asia”, noting that such an expansion is compatible with “the archeological records of eastward expansion of West Asian [peoples].”
The investigation into the descendants of Cush and Phut residing in the Indian sub-continent, revealed that they had migrated from eastern Africa, present day Ethiopia and Somalia, into the Arabian Peninsula and then eastwards into South Asia – refer Chapter XIII India & Pakistan: Cush & Phut. The arrival of non-Indian, Aryan – derived from Iranian – peoples into northern India was either from West Asia, Central Asia or both.
Scientists are correct in stating a. that Haplogroup R1a is a West Eurasian Haplogroup and also in stating that genetic input from ‘Aryan’ migrations has been minimal. Therefore the high diversity and older TMRCA datings of R1a may actually support ancient paternal ancestors outside South Asia, yet who did intermix with Indian and related women and b. in that the majority of Indian Haplogroups by their accumulated micro-satellite variation do attest to an original Haplogroup heritage which clearly preceded the introduction of a lineal R1a descent.
First, what are the original Haplogroups of the Indian males? They are primarily Haplogroup H and secondarily Haplogroup L. Second, when did a DNA infusion from Shem’s line mix with Cush? It is very possible it was during the epoch between the Great Flood and the Tower of Babel, extending from circa 10,837 to 6755 BCE. It was this period which saw the first phase of the great Sumerian (Chapter XXIV Arphaxad & Joktan: Balts, Slavs & the Balkans; and Chapter XVIII Elam & Turkey) civilisation (for it had three) and preceding that, the first Indus Valley civilisation immediately following the flood – Chapter I Noah Antecessor Nulla.
Before continuing with the two specific strains of R1a (Z283 and Z93) in more detail, we will investigate the main sub-Haplogroups of R1a. Forming after R-M207; R2-M479; R1-M173; and beginning from R1a-M420, the next major sub-clades are:
R1a1 M459
R1a1a M17/M198
R1a1a1
M417: a widely found sub-clade, though not in western Europe.

M417 is the major R1a sub-clade from which all the following derive. To assist in reading the Haplogroup letter sequencing after R1a1a1, hyphens are inserted so that mutational descent and progression is more easily followed.
R1a1a1-a
L664/M56: found in Northwest Europe. It is extremely rare with only 1% traced in Sweden, Denmark, Belgium and England. Haplogroup L644 is also located in Western Germany.
R1a1a1-b S224*/M157
R1a1a1-b1 Z283: found within Central and Eastern European men.
R1a1a1-b1a Z282: encompasses most of Eastern Europe.
Especially in Russia at 20% out of 46% of R1a males, as well as in Ukraine and Belarus.
R1a1a1-b1a1 M458: northeastern Europe
R1a1a1-b1a1a L260: West Slavic or Polish (8%) R1a.
Found principally, apart from Poland, in the Czech Republic and Slovakia; as well as in East Germany, Eastern Austria, Slovenia and Hungary. ‘The founding ancestor of R-L260 is estimated to have lived between 2000 and 3000 years ago, i.e. during the Iron Age, with significant population expansion less than 1,500 years ago.’
R1a1a1-b1a2 Z280: includes Finno-Ugric and Balto-Slavic speakers.
Found all over central and eastern Europe, with very low frequencies in the Czech Republic and rarely found in the Balkans.

R1a1a1-b1a3 Z284: Germanic, Scandinavia, Ireland, Scotland and Northern England.
Peaks in Norway with 20% out of 25.5% R1a males.
R1a1a1-b2 Z93: Central Asia, Southwest Asia, South Asia and India.

Ashkenazi Jews carry R1a-CTS6 ‘formed 3,500 years ago’ and Z93 ‘pervaded the genetic pool of the Arabs [R1a-F1345].’ It is also found in the Romani people.
R1a1a1-b2a Z94
R1a1a1-b2a2 M780, L657: India
R1a1a1-c M87, M204, M64.2: very rare found in 1 out of 117 males in southern Iran.
R1a1a1-d P98
R1a1a1-e PK5
R1a1a1-f M434
R1a1a1-g1 (M334 R1a1a1-g1a): only found in one Estonian man.
The highest frequencies of Haplogroup R1a in Europe: ‘Poland (57.5% of the [male] population), Ukraine (40 to 65%), European Russia (45 to 65%), Belarus (51%), Slovakia (42%), Latvia (40%), Lithuania (38%), the Czech Republic (34%), Hungary (32%), Norway (27%), Austria (26%), Croatia (24%), north-east Germany (24%) Sweden (19%), and Romania (18%).’
The origin of R1a lays with Shem and his sons, tracing back through the European Haplogroup lineages: the intermediate Haplogroup I and the older Haplogroup G. The first prime R1a mutation Z283 or R1a1a1-b1 includes the majority of eastern European men and the second prime R1a mutation of Z93 or R1a1a1-b2 includes males outside of Europe, located mainly in Central Asia, Southwest Asia and South Asia. Z93 is a later mutation and its sub-clades show admixture of R1a males into the female line of Cush, the first son of Ham. Though related to Indo-European migrations of Scythians and Indo-Iranians, the initial infusion of R1a as evidenced previously goes back much further in time.

R-Z93 for instance is common in the South Siberian Altai region of Russia (30%+); and in Kyrgyzstan at 6%; while in Iranian populations, it averages 1% to 8%. The most common R1a Haplogroups in Pakistan are M17 and M434. R-M434 (R1a1a1-f) is a sub-clade of Z2125 and was detected in 14 people out of 3,667 people tested. They lived in a restricted geographical range from Pakistan to Oman. ‘This likely reflects a recent mutation event in Pakistan.’ Biblically, Pakistan is Phut the third son of Ham; yet also comprises Ham’s grandson from Mizra, Lehab – refer Chapter XIII India & Pakistan: Cush & Phut. Ethnically, they are a mixed and diverse people.
The main paternal Y-DNA Haplogroups found in Pakistan include:
R1a [37.1%] – J [20.2%] – L [11.6%] – R2 [7.8%] – H [6.2%] – G [6.2%] –
Q [3.4%] – C [3%].
Haplogroups Q and C are men descended in part at least, from Japheth. Haplogroup G is indicative of the Caucasus and a line from Shem. The frequency of R1a is high, in nearly two out of five men and is a conundrum. Particularly when weighed against Haplogroup J1 which is distinctly Arab and equates with the line from Lehab. This leaves Haplogroups H and L. Both these are found in higher frequencies in India; with H highest in Bangladesh (35.7%); and Haplogroup L highest in Sri Lanka (19%).
Which paternal Haplogroup defines the men of Pakistan and consequently Phut? If it is Haplogroup R1a, then how and why did this occur? It should be considered that Shem and Ham both carried the future mutations for Haplogroup R-207 and that R2-479 is Hamitic as it is found principally in Phut and Cush. Nor can Haplogroups H and L be entirely ruled out as secondary Pakistani Haplogroups. The relatively high frequency of Haplogroup J2 stands out as probably the integral marker Haplogroup for Pakistani males.
Eupedia: ‘The Indo-Iranian migrations have resulted in high R1a frequencies in southern Central Asia, Iran and the Indian subcontinent. The highest frequency of R1a (about 65%) is reached in a cluster around Kyrgyzstan, Tajikistan and northern Afghanistan. In India and Pakistan, R1a ranges from 15 to 50% of the population, depending on the region, ethnic group and caste. R1a is generally stronger [in] the North-West of the subcontinent, and weakest in the Dravidian-speaking South (Tamil Nadu, Kerala, Karnataka, Andhra Pradesh) and from Bengal eastward. Over 70% of the Brahmins (highest caste in Hindusim) belong to R1a1, due to a founder effect.’
The Y-DNA Haplogroup frequencies for India overall:
India: R1a [28.3%] – H [23%] – L [17.5%] – R2 [ 9.3%] – J2 [9.1%] –
T [3.1%] – F [3%] – P [2.7%] – C [1.4%] – R1b [0.5%] – Q [0.4%] – G [0.1%]
Indian men possess a fascinating array of paternal Haplogroups. The key Haplogroups identifying Indian men are Haplogroups H, L and possibly R2, which add up to 49.8%. Almost exactly half of all Indian men. The ancient or rare Haplogroups which likely still identify descendants from Cush include, F P and T, totalling 8.9%. Combined they amount to almost 58.7% of Indian men.
Thus the remaining Haplogroups represent admixture from Shem’s line in G, R1a and R1b (total 28.9%) and intermixing from Japheth’s line in C and Q (total 1.8%).
The highest frequencies of Haplogroup H are found in southern India amongst the Dravidians with 32.9% and in Bangladesh higher still with 35.7%. The highest frequency of Haplogroup L is found in Sri Lanka with 19%. In India it is highest in southern India overall, with the Dravidian possessing 11.6%; though the Lambadis carry 17.1% and the Punjabi Indian, 12.1%. Haplogroup R2 is highest in southern India with 21.5% and then eastern India with a frequency of 15.5%.
Haplogroup R1a is found highest in northern India, with the Punjabi carrying 47%, next is Sri Lanka with 27%. This equates to considerable admixture, as India’s position means it has been a thoroughfare for human migration. In other parts of the world, such as Central Asia and southwest Asia the evidence of voluminous migration also reveals an eclectic list of Y-DNA Haplogroups. Thus men from Pakistan who are less mixed from admixture, descending from Phut may only account for the 39.6% represented by Haplogroups J, L and R2. This is intimated in Ezekiel 30:5.
Eupedia: ‘Maternal lineages in South Asia are, however, overwhelmingly pre-Indo-European. For instance, India has over 75% of “native” mtDNA M and R lineages and 10% of East Asian lineages. In the residual 15% of haplogroups, approximately half are of Middle Eastern origin. Only about 7 or 8% could be of “Russian” (Pontic-Caspian steppe) origin, mostly in the form of haplogroup U2 and W (although the origin of U2 is still debated). European mtDNA lineages are much more common in Central Asia though, and even in Afghanistan and northern Pakistan. This suggests that the Indo-European invasion of India was conducted mostly by men through war. The first major settlement of Indo-Aryan women was in northern Pakistan, western India (Punjab to Gujarat) and northern India (Uttar Pradesh), where haplogroups U2 and W are the most common today.’
This paints a very different picture compared with the Y-DNA Haplogroup frequencies and reveals the extent of foreign male admixture into the South Asian gene pool.
Eupedia: ‘Comparing the regions where haplogroup R1a is found today with the modern mtDNA frequencies, it transpires that the maternal lineages that correlate the most with Y-haplogroup R1a are mt-haplogroups C4a, H1b, H1c, H2a1, H6, H7, H11, T1a1a1, U2e, U4, U5a1a and W, as well as some subclades of I, J, K, T2 and V.’
Both Haplogroups R1a and R1b are associated with the ‘diffusion of the A111T mutation of the SLC24A5 gene, which explains approximately 35% of skin tone difference between Europeans and Africans, and most variations within South Asia. The distribution pattern of the A111T allele (rs1426654) matches almost perfectly the spread of Indo-European R1a and R1b lineages around Europe, the Middle East, Central Asia and South Asia. R1a populations have an equally high incidence of this allele as R1b populations. On the other hand, the A111T mutation was absent from the [ancient] R* sample (Mal’ta boy) from Siberia, and is absent from most modern R2 populations in Southeast India… Consequently, it can be safely assumed that the mutation arose among the R1* lineage…’ and that R2 is a distinct Hamitic line separate from R1.
Famous male R1a individuals include: Francis Drake, L664; Somerled of Argyll, founder of Clan Somhairle, father of the founder of Clan MacDougall and the paternal grandfather of the founder of Clan Donald [which includes certain MacDonalds and MacAlisters], L448/L176.1 from Z284* (Germanic, Scandinavian, British), though not all MacDonalds, MacAlisters and MacDougalls descend from Somerled as the 70% majority are members of the Celtic R1b Haplogroup; Clan Cochrane and Earl of Dundonald, L448; Clan Home or Hume, L448; David Hume, L448; Tom Hanks*, R1a-Z284; Nikola Tesla, R1a-M458 (L1029).

Tom Hanks ancestry is of interest as while his mother was Portuguese, his father had English ancestry and through his line, Hanks is a distant cousin of both Nancy Hanks and her son President Abraham Lincoln.

Even so, his Y-DNA Haplogroup R1a-Z284 belies a paternal ancestor with an Eastern European Haplogroup and a Scandinavian mutation through intermixing. Remember, an unmixed English ancestry would carry R1b-U106 or downstream mutations.
Rudolph Hess: “The DNA analysis of the only known extant DNA sample from prisoner ‘Spandau #7’ proved to be a match to the Hess male line, thereby refuting the Doppelgänger Theory”, M458 (Proto-Slavic). Max von Sydow, Z280, (P269); King Willem-Alexander of the Netherlands, Z280 (S18681); Alexander Pushkin, Russian Poet, Z92; the Ottoman Dynasty: ‘All sultans of the Ottoman Empire (1299-1922) descend in patrilineal line from Osman I, making it one of the longest reigning Y-chromosomal lineage in history’, Z93; Benjamin Netanyahu, Z93 (Y2630 Jewish sub-clade); Jesse James*, ‘R1a-Y2395 > Z284> L448 > CTS4179> YP386, a Scandinavian branch of R1a also found in Britain.’
The final Y-DNA Haplogroup is R1b (M343, M415). It is a ‘younger’ sibling to R1a (M420). Both descend from R1 (M173) and R-M207 (K2b2a2). The Haplogroup R branch with the older Q -M242 (K2b2a1), derives from P1-M45 (K2b2a). P1 is the primary branch from P-295 (K2b2) and P descends from K2b (P331).

Encyclopaedia: ‘Haplogroup R1b (R-M343), [was ] previously known as Hg1 and Eu18… It is the most frequently occurring paternal lineage in Western Europe, as well as [in] some parts of Russia (e.g. the Bashkirs) [Bashkir men have also been found to belong to R1b-U152, while some from southeastern Bashkortostan, are Haplogroup Q-M25 (Q1a1b) as opposed to R1b-M73 which is found in 23.4% of males] and pockets of Central Africa (e.g. parts of Chad and among the Chadic-speaking minority ethnic groups of Cameroon).
The clade is also present at lower frequencies throughout Eastern Europe, Western Asia, as well as parts of North Africa, South Asia and Central Asia. The age of R1 was estimated by Tatiana Karafet et al. (2008) at between 12,500 and 25,700 BP, and most probably occurred about 18,500 years ago. Since the earliest known example has been dated at circa 14,000 BP, and belongs to R1b1 (R-L754)… R1b must have arisen relatively soon after the emergence of R1.’
The emergence of Haplogroup R would have been post-flood – which occurred in 10,837 BCE according to an unconventional chronology – and within 12,000 years ago amongst the descendants of Shem. All Haplogroup mutations have arisen in a concertinaed fashion, without tens of thousands of years between them but rather thousands to hundreds of years.

R1b has two primary branches: R1b1 (L278) and R1b2 (M335, PH155). R1b2 is very rare and has been found in Bahrain, India, Nepal, Bhutan, Tajikistan, Turkey and even western China. This clade R1b2 is a residue of ancient R1b inhabitants in these regions and not an evolved R1b mutation. From L278 is L754 (R1b1-a) and from L745 are two major divisions with L388 (R1b1-a1) and V88, or R1b1-a2 (M18, V35, V69). Haplogroup R1b1a2 is found rarely, either in the Levant or more commonly in North Central Africa; though M18 and V35 are found almost exclusively on the Italian Island of Sardinia. The maternal lineages associated with the spread of V88 in Africa, include mtDNA Haplogroups: J1b, U5 and V.
From L388 is P297 (R1b1-a1a). Here it divides into R1b1-a1a1 (M73, M478) a rare clade found in Central Asia, the Caucasus, Siberia and Mongolia, and the major sub-clade R1b1-a1a2 or M269, widespread throughout western Europe – re-classified as R1b1a1b since 2018. Both V88 and M73 are residue Haplogroup clades from ancient R1b male inhabitants and integration with local women.
Encyclopaedia:
‘Early human remains found to carry R1b include:
- Several males of the Iron Gates Mesolithic in the Balkans buried between 11200 and 8200 BP carried R1b1a1a [P297].
- Several males of the Mesolithic Kunda culture and Neolithic Narva culture buried in the Zvejnieki burial ground in modern-day Latvia c. 9500–6000 BP carried R1b1b.
- Several Mesolithic and Neolithic males buried at Deriivka and Vasil’evka in modern-day Ukraine c. 9500-7000 BP carried R1b1a [L745].
- A male of the Botai culture in Central Asia buried c. 5500 BP carried R1b1a1[a1] (R1b-M478).
No confirmed cases of R1b* (R-M343*)… have been reported in peer-reviewed literature. In early research, because R-M269, R-M73 and R-V88 are by far the most common forms of R1b, examples of R1b (xM73, xM269) were sometimes assumed to signify basal examples of “R1b*”. However, while the paragroup R-M343 (xM73, M269, V88) is rare, it does not preclude membership of rare and/or subsequently-discovered, relatively basal subclades of R1b such as R-L278*… R-P297*… or R-PH155… The population believed to have the highest proportion of R-M343 (xM73, M269, V88) are the Kurds of southeastern Kazakhstan with 13%.
R-L278 among modern men falls into the R-L754 and R-PH155 subclades, though it is possible some very rare R-L278* may exist as not all examples have been tested for both branches. Examples may also exist in ancient DNA, though due to poor quality it is often impossible to tell whether or not the ancients carried the mutations that define subclades. R-L754 contains the vast majority of R1b. The only known example of R-L754* (xL389, V88) is also the earliest known individual to carry R1b: “Villabruna 1”, who lived circa 14,000 years BP (north east Italy).
R-L389, also known as R1b1a[1] (L388/PF6468, L389/PF6531), contains the very common subclade R-P297 and the rare subclade R-V1636. It is unknown whether all previously reported R-L389* (xP297) belong to R-V1636 or not. The SNP marker P297 was recognised in 2008 as ancestral to the significant subclades M73 and M269, combining them into one cluster. A majority of Eurasian R1b falls within this subclade, representing a very large modern population. Although P297… has not yet been much tested for [itself], the same population has been relatively well studied in terms of other markers.’
R1b-M269 is the most common R1b Haplogroup – carried by some ‘110 million males in Europe’ – and the defining marker for Shem’s descendants aside from R1a and not withstanding those men who carry Haplogroups G2a, I1 and I2a1. R1b-M269 ‘is closely associated with the diffusion of Indo-European languages…’
Encyclopaedia: ‘Distribution of R-M269 in Europe increases in frequency from east to west. It peaks at the national level in Wales at a rate of 92%, at 82% in Ireland, 70% in Scotland, 68% in Spain, 60% in France… about 60% in Portugal, 50% in Germany… 47% in Italy, 45% in Eastern England and 42% in Iceland. R-M269 reaches levels as high as 95% in parts of Ireland.’
M269 (R1b1a-1a2) diverges into a number of significant sub-clades. Joint oldest being PF7562 (R1b1a-1a2b), re-classified as R1b1a1b2 and located in the Balkans, Turkey and Armenia; and L23 (R1b1a1a-2a), re-classified as R1b1a1b1. Related to PF7562 is one of two branches from L23; Z2103 (R1b1a-2a2) found in Eastern Europe and West Asia. Sub-clades within L23 ‘appear to be found at their highest frequency in the Central Balkans, especially Kosovo with 7.9%, North Macedonia 5.1% and Serbia 4.4%.’
The other is L51 or R1b1a-1a2a1 (M412), indicative of Central Europe and also found in southern France and northern Italy R-L51*/R-M412*. Though, deriving from L51 is L151 ([P310, P311] R1b1a-1a2a1a) and its sub-clades which ‘include most males with R1b in Western Europe. The oldest samples classified as belonging to R-M269, have been found in Eastern Europe and [the] Pontic-Caspian steppe, not Western Asia. Western European populations are divided between the R-P312/S116 and R-U106/S21 subclades of R-M412 (R-L51).’
R1b-Z2103
Eupedia: ‘Haak et al. (2015) tested six Y-DNA samples from… the Volga-Ural region, and all of them turned out to belong to haplogroup R1b. Four of them were positive for the Z2103 mutation. In all likelihood, R1b-Z2103 was a major lineage of the Poltavka culture, which succeeded to the Yamna culture between the Volga River and the Ural mountains. It eventually merged with the Abashevo culture (presumably belonging chiefly to R1a-Z93) to form the Sintashta culture. Through a founder effect or through political domination, R1a-Z93 lineages would have outnumbered R1b-Z2103 after the expansion to Central and South Asia… R1b-Z2103 would have become an Indo-Iranian lineage like R1a-Z93. This is true of two Z2103 subclades in particular: L277.1 and L584. The former is found in Russia to Central Asia then to India and the Middle East, just like the R1a-L657 subclade of Z93.’
Z2103 is an ancient R1b clade which perhaps hasn’t mutationally evolved as far and so is not found in modern western European populations. Showing it is likely a R1b residue from ancient admixture, as is the case with M335, V88, M73 and PF7562. The alternative explanation is that Z2103 is primarily indicative of Elam’s descendants, the Turks – Chapter XVIII Elam & Turkey.
Distribution (below) for Haplogroup R1b-ht35 (Z2103) in Europe

Descending from L151 are three R1b lineages via L11 ([P310] R1b1a-2a1a):
U106 ([S21] R1b1a–1a2a1a1), P312 ([S116] R1b1a-1a2a1a2); and CT4528 (R1b1a-1a2a1a3a).
R1b-U106
L11 is found in Central England, though the first major R1b lineage from L151 is U106 (S21, M404) which encompasses all the Germanic speaking nations of northwestern Europe; including: Germany (Ishmael), Austria (Hagar), Switzerland (Haran), Scandinavia (Keturah) the Benelux nations (Keturah), the United Kingdom (Judah, Benjamin, Simeon, Reuben) and Ireland (Gad) – bold = Abraham.
‘It appears to represent over 25% of R1b in Europe. In terms of percentage of total population, its epicenter is Friesland, where it makes up 44% of the population. In terms of total population numbers, its epicenter is Central Europe, where it comprises 60% of R1 combined.’

One cannot but equate U106 with the Patriarch Abraham. With the exception of Austrians who derive from Hagar and her mystery husband and the Swiss who descend from Abraham’s brother Haran, these nations embody the progeny of Abraham; either through Ishmael, Isaac or the sons of Keturah – refer Chapter XXVI The French & Swiss: Moab, Ammon & Haran; Chapter XXVII Abraham & Keturah – Benelux & Scandinavia; and Chapter XXVIII The True Identity & Origin of Germans & Austrians -Ishmael & Hagar. Granted, the beginning of the U106 mutation may not have originated with Abraham, though the compelling evidence is that this sub-Haplogroup identifies predominantly with his lineage.
Significant branches deriving from U106 include: FGC3861 (R1b1a1a-2a1a1a); Z18 (R1b1a1a-2a1a1b); Z381 (S263); FGC396 (R1b1a1a-2a1a1d); and S12025 (R1b1a1a-2a1a1e).
Sub-clade Z381 has three main offshoots: S264 ([Z156] R1b1a1a–2a1a1c1); S499 ([Z301] R1b1a1–2a1a1c2); and M1994 (R1b1a1a-2a1a1c3).
Also stemming from Z381 is M323 found uniquely in Britain. Significant branches of Z301 include L48 and S1688 from which U198 derives. Sub-clade U198 is common throughout Southern and Eastern England.

Eupedia: ‘The principal Proto-Germanic branch of the Indo-European family tree is R1b-S21 [U106, M405]… This haplogroup is found at high concentrations in the Netherlands and north-west Germany. It is likely that R1b-S21 lineages expanded in this region through a founder effect during the Unetice period, then penetrated into Scandinavia around 1700 BCE (probably alongside R1a-L664), thus creating a new culture, that of the Nordic Bronze Age (1700-500 BCE). R1b-S21 would then have blended for more than a millennium with preexisting Scandinavian populations, represented by haplogroups I1, I2-L801, R1a-Z284. When the Germanic Iron Age started c. 500 BCE, the Scandinavian population had developed a truly Germanic culture and language, but was divided in many tribes with varying levels of each haplogroup. R1b-S21 became the dominant haplogroup among the West Germanic tribes, but remained in the minority against I1 and R1a in East Germanic and Nordic tribes…
The presence of R1b-S21 in other parts of Europe can be attributed almost exclusively to the Germanic migrations that took place between the 3rd and the 10th century. The Frisians and Anglo-Saxons [and the Jutes] disseminated this haplogroup to England and the Scottish Lowlands, the Franks to Belgium and France, the Burgundians to eastern France, the Suebi to Galicia and northern Portugal, and the Lombards to Austria and Italy. The Goths help propagate S21 around Eastern Europe, but apparently their Germanic lineages were progressively diluted by blending with Slavic and Balkanic populations… Later the Danish and Norwegian Vikings have also contributed to the diffusion of R1b-S21 (alongside I1, and R1a)… mainly in Iceland, in the British Isles, [and] in Normandy…
From the Late Middle Ages until the early 20th century, the Germans expanded across much of modern Poland, pushing as far as Latvia to the north-east and Romania to the south-east. During the same period the Austrians built an empire comprising what is now the Czech Republic, Slovakia, Hungary, Slovenia, Croatia, Serbia, and parts of Romania, western Ukraine and southern Poland. Many centuries of German and Austrian influence in central and Eastern Europe resulted in a small percentage of Germanic lineages being found among modern populations.
O’Sullivan et al. (2018) tested the genomes of Merovingian nobles from an early Medieval Alemannic graveyard in Baden-Württemberg’ – refer Chapter XXXIV Dan: The Invisible Tribe. ‘Apart from one individual belonging to haplogroup G2a2b1, all men were members of R1b, and all samples that yielded deep clade results fell under the R1b-U106 > Z381 > Z301 > L48 > Z9 > Z325 clade’ – see Phylogenetic tree above.
The lineage of the Kings of France was inferred from the Y-DNA of several descendant branches… and also belongs to R1b-U106 > Z381′ – refer Chapter XXVI The French & Swiss: Moab, Ammon & Haran. Their earliest-known male-line ancestor was from Robert II, Count of Hesbaye, a Frankish nobleman from present-day Belgium.
The House of Wettin… one of the oldest dynasties in Europe, which ruled over many states at various times in history, was yet another well-known noble Germanic lineage part of R1b-U106 > Z381′ – refer Chapter XXIX Esau: The Thirteenth Tribe; and Chapter XXX Judah & Benjamin – the Regal Tribes.
Distribution (below) for Haplogroup R1b-S21 (U106) in Europe
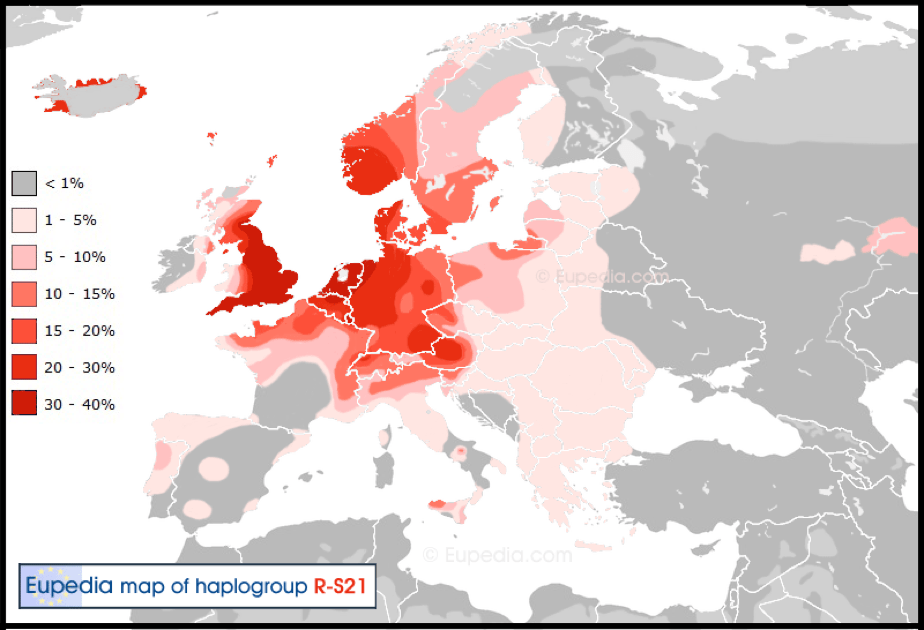
The second dominant lineage from L11, apart from the Germanic U106, is R1b-S116 (P312). It has three prominent downstream groups: DF27 (S250); U152 (S28); and L21 (M529). Haplogroup L21 is classified as Atlantic-Celtic; U152 as Italian-Gallic; and DF27 as Iberian-Atlantic. ‘Myres et al. described [R-P312] as originating in and spreading from the west of the Rhine basin. R-P312 has been the subject of significant, ongoing study concerning its complex internal structure.’
This raises an important point, for this writer remains unconvinced in the exact thread of the R1b genetic tree at its tail end – that is, it’s most recent mutations. Input from geneticists would be welcome concerning this idea. Briefly, the Atlantic Celtic M529 would seem logically to be either next to the Proto-Germanic U106 (beneath L11) or deriving from U106. Similarly, the Italo-Gaulish U152 would also seem better placed deriving from L11 and located between the Proto-Germanic U106 and (the Ibero-Atlantic DF27 stemming from) P312.
R1b-DF27
Two distinct lines from DF27 include the ZZ12 phylogenetic tree and Z195.

Haplogroups of note from the Z195 tree include M153 which is found mostly in Basques and Gascons, though also amongst Iberians in general. R1b-M167 is relatively common among Basques (11%) and Catalans (22%). It is also found in the Spanish, French, Germans and British, principally in Cornwall and Wales. The sub-clade L165 ‘is defined by the presence of the marker S68… It is found in England, Scandinavia, and Scotland… [and] mostly found in the Northern Isles and Outer Hebrides…’
Eupedia: ‘Martiniano et al. (2017) sequenced the genomes of various skeletons from West Iberia dating from the Middle and Late Neolithic, Chalcolithic and Middle Bronze Age (since the Early Bronze Age did not reach that region). They found that Neolithic and Chalcolithic individuals belonged to Y-haplogroups I*, I2a1 and G2a. In contrast, all three Bronze Age Portuguese men tested belonged to R1b (one M269 and two P312)… they carried Neolithic Iberian maternal lineages (H1, U5b3, X2b)…’
Though DF27 is found in western Europe it is primarily indicative of the Spanish Visigoths and Portuguese Suebi, the descendants of Shem’s fifth son Aram – refer Chapter XXIII Aram & Tyre: Spain, Portugal & Brazil; and Chapter XV The Philistines: Latino-Hispano America.
Distribution (below) for Haplogroup R1b-DF27 (S250) in Europe
R1b-U152
Encyclopaedia: ‘R-U152 is defined by the presence of the marker U152, also called S28. Its discovery was announced in 2005 by EthnoAncestry and subsequently identified independently by Sims et al. (2007). Myres et al. report this clade “is most frequent (20–44%) in Switzerland [Haran], Italy [Nahor], France [Lot] and Western Poland, with additional instances exceeding 15% in some regions of England and Germany.” Similarly Cruciani et al. (2010) reported frequency peaks in Northern and Central Italy and France. Out of a sample of 135 men in Tyrol, Austria, 9 tested positive for U152/S28. King et al. (2014) reported four living descendants of Henry Somerset, 5th Duke of Beaufort^ in the male line tested positive for U-152.’
R1b-U152 while found in Central Europe, is indicative of the Northern and Central Italians, the Swiss and the French: equaling the descendants of Abraham’s two older brothers Nahor, Haran and his nephew Lot respectively.
Eupedia: ‘Furtwangler et al. (2020) analysed 96 ancient genomes from Switzerland, Southern Germany, and the Alsace region in France, covering the Middle/Late Neolithic to Early Bronze Age. They confirmed that R1b arrived in the region during the transitory Bell Beaker period (2800-1800 BCE). The vast majority of Bell Beaker R1b samples belonged to the U152 > L2 clade (11 out of 14; the other being P312 or L51).’

‘Antonio et al. (2019) analysed the genomes of Iron Age Latins dating between 900 and 200 BCE, and the samples tested belonged primarily to haplogroup R1b-U152 (including the clades L2, Z56 and Z193), as well as one R1b-Z2103 and one R1b-Z2118.’
Distribution (below) for Haplogroup R1b-S28 (U152) in Europe
R1b-L21
Haplogroup R1b-L21 is also known as M529 and S145. It is a quintessentially Celtic group, though it is also found in England. ‘Myres et al. report it is most common in Ireland [Gad, Reuben], Scotland [Benjamin] and Wales [Simeon]’ accounting for between 25% to 50% of the whole male population. Haplogroup M529 (R1b-L21) with U198 and M323 from R1b-U106 are prime groups reflecting the descendants of Jacob’s sons and the Celtic-Saxon-Viking peoples.

Eupedia: ‘The Proto-Italo-Celto-Germanic R1b people… [as the] first wave of R1b presumably carried R1b-L21 lineages in great number (perhaps because of a founder effect), as these are found everywhere in western, northern and Central Europe. Cassidy et.al (2015) confirmed the presence of R1b-L21 (DF13 and DF21 subclades) in Ireland around 2000 BCE. Those genomes… differed greatly from the earlier Neolithic Irish samples. This confirms that a direct migration of R1b-L21… was responsible for the introduction of the Bronze Age to Ireland.
The early split of L21 from the main Proto-Celtic branch around Germany would explain why the Q-Celtic languages (Goidelic and Hispano-Celtic) diverged so much from the P-Celtic branch (La Tène, Gaulish, Brythonic)… Some L21 lineages from the Netherlands and northern Germany later entered Scandinavia with the dominant subclade of the region, R1b-S21/U106. The stronger presence of L21 in Norway and Iceland can be attributed to the Norwegian Vikings, who had colonised parts of Scotland and Ireland and taken slaves among the native Celtic populations, whom they brought to their new colony of Iceland and back to Norway… about 20% of all Icelandic male lineages are R1b-L21 of Scottish or Irish origin.
In France, R1b-L21 is mainly present in historical Brittany and in Lower Normandy. This region was repopulated by massive immigration of insular Britons in the 5th century due to pressure from the invading Anglo-Saxons. However, it is possible that L21 was present in Armorica [earlier]… given that the tribes of the Armorican Confederation of ancient Gaul already had a distinct identity from the other Gauls and had maintained close ties with the British Isles…’
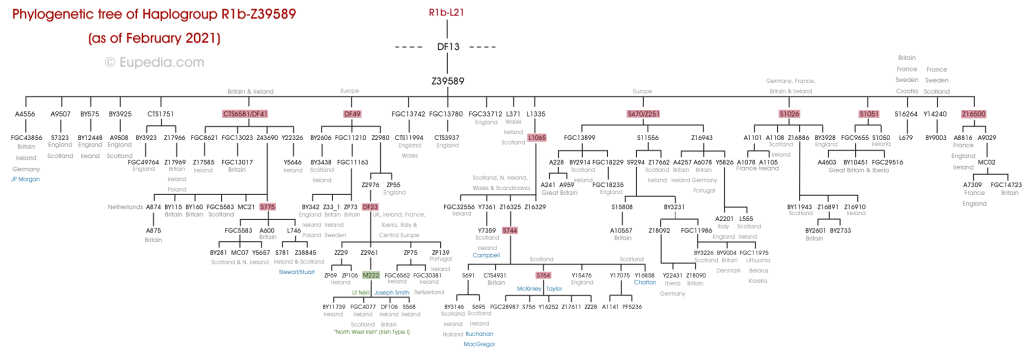
R1b-L21 sub-clades of interest include: R-M222; L159.2; L193; L226; and L371.
Distribution (below) for Haplogroup R1b-L21 (M529) in Europe
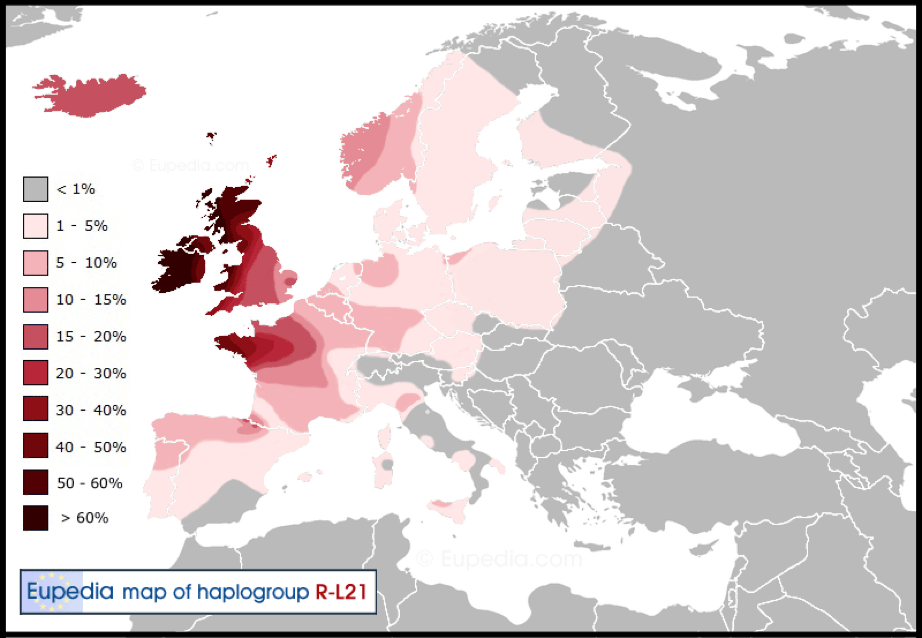
Encyclopaedia: ‘… [M222] within R-L21 is… estimated to have arisen between 1400 and 2000 BCE. It is particularly associated with male lines which are Gaelic (Irish or Scottish), but especially north-western Irish… it is suggested to have been the Y-chromosome haplogroup of the Ui Neill dynastic kindred of ancient Ireland, often referred to as that of the prominent Dark Age monarch Niall of the Nine Hostages… [also] associated with the closely related Connachta dynasties, the Ui Bruin and Ui Fiachrach. M222 is also found as a substantial proportion of the population of Scotland which may indicate substantial settlement from northern Ireland or at least links to it. Those areas settled by large numbers of Irish and Scottish emigrants such as North America have a substantial percentage of M222’ – refer Chapter XXXIV Dan: The Invisible Tribe.
The L159.2 sub-clade within R-L21 is known ‘as… a parallel mutation that exists inside haplogroup I2a1 (L159.1). L159.2 appears to be associated with the Kings of Leinster and Diarmait Mac Muurchada; Irish Gaels belonging to the Laigan. It can be found in the coastal areas of the Irish Sea including the Isle of Man and the Hebrides, as well as Norway, western and southern Scotland, northern and southern England, northwest France, and northern Denmark. Many surnames with [the L193]… marker are associated geographically with the western “Border Region” of Scotland. A few other surnames have a Highland association. R-L193 is a relatively young subclade likely born within the last 2000 years… marker L226, also known as S168. Commonly referred to as Irish Type III, it is concentrated in central western Ireland and associated with the Dal gCais kindred… marker L371, [is] referred to as the Welsh modal and associated with ancient Welsh Kings and Princes.’
Prominent members of R1b include: Charles Darwin; Kevin Bacon; Robert Downey Jr; Harry Connick Jr. ‘Yehia Z Gad… at the Ancient DNA lab of the National Museum of Egyptian Civilization in Cairo retrieved the DNA of several members of the 18th Dynasty of Egypt… which included Amenhotep I to III, Thutmose I to IV… Akhenaten and Tutankhamun. The Y-DNA analysis established that the royal male lineage belonged to Y-haplogroup R1b.’
Rogaev [2009] ‘tested the DNA of the presumed grave of Tsar Nicolas II of Russia and all his five children, and compared them against archival blood specimens from Nicholas II as well as against samples from descendants of both paternal and maternal lineages. The results unequivocally confirmed that the grave was the one of the last Russian Royal family’ – Chapter XX Will the Real Assyria Stand Up: Asshur & Russia. ‘Nicholas II belonged to Y-haplogroup R1b and mt-haplogroup T2‘ – Chapter XXIX Esau: The Thirteenth Tribe.
‘Consequently, all Russian emperors of the Romanov dynasty since Peter III (1728-1762) also belonged to haplogroup R1b [particularly the later Tzars of the House of Romanov who descended from the ‘House of Holstein-Gottorp in Schleswig-Holstein’]. This paternal lineage ultimately descends from the House of Oldenburg, which includes all the Kings of Denmark since Christian I (reigned from 1448) as well as several Kings of Norway, Sweden and Greece, and the current heirs to the British throne’ Prince William and his son Prince George – Article: The Life & Death of Charles III.
John Adams, second President and his son John Quincy Adams, sixth president, R1b-S2100; Thomas Edison, R1b-S2100; Clan Grant, R1b-P312 (DF19); Clan Armstrong, astronaut Neil Armstrong, P312; Woodrow Wilson, 28th President, P312; Nicolaus Copernicus, Renaissance astronomer, R1b-P310 (and mtDNA H).
Clan Bruce, Robert the Bruce and David II of Scotland and High King of Ireland, Edward Bruce, Earls of Elgin and Earls of Kincardine, R1b-DF27 > ZZ12 > Z46512 > FGC78762 > ZZ41 > S7432; Clan Boyle, Earls of Glasgow, DF27 (Z196); House of Bernadotte, Royal House of Sweden since 1818, DF27 (Z195).
Matthew Calbraith Perry, ‘the man who forced Japan to open its ports to western ships’, DF27 (Z196); George W Bush, 41st President, descended from Reynold Bush (1600-1686) who emigrated from Fering Parish in Essex England, to the Massachusetts colony about 1640, R1b-DF27 > Z196 > Z209 > CTS4065 > S16864; Pierre Trudeau and his son Justin Trudeau, DF27 (Z196).
Clan Murray and Clan Sutherland “both descend from a Flemish nobleman by the name of Freskin, who settled in Scotland during the reign of King David I and was granted lands in West Lothian and the ancient Pictish kingdom of Moray (which would become known as Sutherland). Freskin’s descendants were designated by the surname de Moravia (“of Moray” in the Norman language), which later became ‘Murray’. Freskin’s great-grand-son was William de Moravia (c. 1210–1248).. [and] became 1st Earl of Sutherland, a title that the clan chief [kept] until 1535, when it passed to Clan Gordon. Clan Murray descends from William’s cousin… the ancestral Sutherland line belongs to R1b-DF27 > ZZ12 > FGC23071 > FGC23066 > BY48361 > BY130907 > BY67446 and has Y-chromosomal matches in modern Flanders, confirming Freskin’s origins. It is believed that Clan Douglas also descends from Freskin and… indeed matches the Sutherland and Murray haplotype.”
Before moving on to the next R1b sub-Haplogroup, it is important to highlight that even though these illustrious men are principally British through and through, they all without exception had at one time a paternal ancestor who descended from an R1b line which was not of Abraham (U106), but from Aram (DF27) and his four sons; equating today to the peoples of Spain and Portugal and their Spanish and Portuguese descendants in the Americas.
House of Hapsburg, R1b-U152 > L2 > Z41150 > DF90 > FGC59564; Richard III of England ‘… three modern relatives with the surname Somerset and descended from House of Lancaster all belonged to haplogroup R1b-U152 (x L2, Z36, Z56, M160, M126 and Z192). Although this points to a non-paternity at some time in the Plantagenet lineage, it is likely that most if not all Dukes of Beaufort, and possibly most Plantagenets monarchs outside the House of York belonged to R1b-U152.’ Clan Erskine, U152 (Z36); Grover Cleveland, 22nd and 24th President, L2 (L20); Kevin Costner, L2; Matthew Perry, L2 (Z142).
The same situation exists for U152 as it does with DF27. In this case the above men have had a paternal ancestor descended not from Abraham, but one of his brothers; either Nahor, northern and central Italians; or Haran, Switzerland and by his son Lot, the French and French Canadians.
So it is of great interest that Abraham Lincoln who possessed a rare mtDNA Haplogroup from his mother (X1c) should have inherited R1b-U152 from his father.

Abraham Lincoln’s Mother, Nancy Hanks
Recall, Haplogroup X is found in only 1% of the world’s population; with X1c being rarer still. It has been found in Norway, Ireland and interestingly, Italy.

Abraham Lincoln^, 16th President, likely belonged to ‘R1b-U152 > L2 > Z142 > Z150 > S20376… as several descendants from Samuel “the weaver” Lincoln, who was Abraham Lincoln’s great-great-great-great-grandfather… all [shared] the same haplotype.’
Perhaps this explains Lincoln’s features and colouring as not being an archetypal Celtic-Saxon-Viking lineage. That said, the sub-clade of U152 which Abraham Lincoln perhaps possessed is a mutation specifically carried by British men (Z150) and in found in England (S20376).

Though a further explanation is found in the article, The Establishment: Who are they… What do the want? Where it is offered that a. Nancy Hanks was of Scottish extraction and b. Abraham Lincoln’s biological father was not actually Thomas Lincoln (whom in all honesty he does not resemble) and was in fact his adoptive father.
Abraham Lincoln’s real Father was allegedly A A Springs, who originally came from a line of the Rothschilds which had changed their name. Thus Lincoln’s Y-DNA Haplogroup could have been a different R1b and even R1a, J2, J1 or E1b1b.

A A Springs and Abraham Lincoln
Recall that Tom Hanks (R1a) is related to Abraham Lincoln on Hank’s father’s side.
O’Neil Dynasty, Gaelic Irish lineage, Northern Ireland, descended from Niall of the Nine Hostages, R1b-L21 (DF13, DF49); Clan Maclean, L21 (DF13, DF1); Clan Gregor (McGregor), folk hero Rob Roy MacGregor, L21 (DF13); Clan Campbell, L21 (DF13); House of Stuart, ‘who ruled Scotland from 1371, then also England and Ireland from 1603 until 1707, belongs to R1b-L21 > DF13 > Z39589 > DF41/S524 > Z43690 > S775 > L746 > S781. The most prominent members were King Robert II of Scotland, Kings James I, Charles I, Charles II and James II of England & Ireland.’
Clan MacKenzie, R1b-L21 (DF13); George Washington, 1st President, R1b-L21 > DF13 > ZZ10 > Z253 > Z2186 > BY2744; Zachary Taylor; L21 (DF13); William Gladstone, L21 (DF13); Rutherford B Hayes, 19th president, L21 (DF13); J P Morgan, financier and banker, L21 (DF13); William McKinley, 25th President, L21 (DF13); Warren G harding, 29th President, L21 (DF13); Che Guevara, Argentine Marxist revolutionary, R1b-L21; Matt LeBlanc, L21 (DF63).
Notice the royal lineage of the Stuart kings and numerous American presidents all falling under the Celtic R1b Haplogroup M529. This sub-clade with the Germanic U106, are the defining markers for men descended from Isaac, the son of Abraham.
John Smith, ‘the founder of Mormonism and the Latter Day Saint movement, belonged to haplogroup R1b-M222 (R1b-L21 > DF13 > DF49 > Z2980 > Z2976 > DF23 > Z2961 > S645 > Z2965 > M222)’ – refer Chapter XXXIV Dan: The Invisible Tribe. The following personalities are all members of M222: Henry Louis Gates, American writer; Bill O’Reilly, American television host; Bill Maher, American comedian; Rory Bremner, Scottish comedian; Adrian Grenier, American actor.
Clan Boyd, Earl of Kilmarnock, R1b-U106 > Z381 > S1684 > U198 > S15627 > DF89 > FGC12770 > FT69836 > JFS0024 (the sub-clade U198 is typically an English haplotype); Franklin Pierce, 14th President, R1b-U106 > Z381 > Z156 > S497 > DF96; Alec Baldwin, Z381; Woody Harrelson, Z18; Clan Gordon, R1b-U106; Benjamin Franklin, R1b-U106 (Z18, DF95); James K Polk, 11th President U106 (S263, L48 [Z301]); Ulysses S Grant, 18th president, L48 (Z301); Ernest Hemingway, L48 (Z301).
House of Bourbon, R1b-U106 (Z381). ‘All kings of France being descended in patrilineal line from Robert the Strong (820-866), unless a non-paternity event happened some time before Louis XIII… belonged to the same R1b-Z381 lineage. The House of Bourbon also includes all the kings of Spain from Philip V (1683-1746) to this day with King Juan Carlos, all the kings of the Two Sicilies, the grand dukes of Luxembourg since 1964, and of course all the dukes of Orléans and the dukes of Bourbon.’

‘The lineage of the House of Wettin was identified as R1b-U106 > Z2265 > Z381 > Z156 > Z305 > Z307 > Z304 > DF98 > S18823 > S22069 > Y17440 > A6535… Members of the House of Wettin include the Kings Edward VII, George V, Edward VIII and George VI of the United Kingdom, all the Kings of the Belgians, the Kings of Portugal from 1853 to 1910, the Kings of Bulgaria from 1887 to 1946, several Kings of Poland and Grand Dukes of Lithuania, the Margraves of Meissen from 1075 to 1423, the Electors of Saxony from 1423 to 1806, the Kings of Saxony from 1806 to 1918, and the rulers of the numerous smaller Saxon duchies.’

The House of Wettin haplotype S8350 while loosely British, is more accurately ‘Germanic’ as in Germany rather than England – refer Chapter XXX Judah & Benjamin – the Regal Tribes; and Chapter XXIX Esau: The Thirteenth Tribe.
Thomas Cecil, 1st Earl of Exeter, R1b-U106 > Z381 > L48 > Z9 > Z331 > Z330 > Y6669 > S21728 > FGC18850 > Y21406 > Y20959 > FGC51954.
Of interest to this writer is the Z30 sub-clade downstream from Z301, which includes: Clan Sinclair, Earl of Orkney and Earl of Caithness, R1b-U106 > Z381 > L48 > Z9 > Z30 > Z7 > Z346 > S5246 > S5629 > FGC15254 > FGC35613 > ZS5151; William Howard Taft (Skull & Bones), 27th President, R1b-U106 > Z381 > Z301 > Z30 > Z338 > FGC1954; and James D Watson, one of the first two ‘human beings to have their whole genome sequenced… and [co-discoverer] of the structure of DNA… [member] of Y-DNA haplogroup R1b-S21 (U106)… subclades… L48 > Z9 > Z30′ a descendent of Scottish ancestors.
Nathan Bedford Forrest (1821-1877) ‘was a prominent Confederate Army general during the American Civil War, renowned as a cavalry leader and military strategist.’

‘He was the only general on either side who began as a private. After the war he became the first Grand Wizard of the Ku Klux Klan, which has made him a controversial figure in American history, R1b-U106 > S263 > S499 > L48 > Z9 > Z30 > Z349 > Z2 > Z7 > S5945 > FGC17344 > Y28576 > FGC51332.’

From our previous summary of Y-DNA Haplogroups A to I, we have in addition, J through to T. Recall Haplogroups A, B, E1a, E1b1a and E2 are associated with peoples of Black African heritage. Haplogroup E1b1b is primarily associated with Berbers in North Africa and related ‘non-Arab’ peoples in southern Europe.
Added now to these peoples are the intermediate mutations of J1 and J2, found in – related peoples through admixture in southern Europe and – origination with men now in West Asia and the Arabian Peninsula.
Haplogroup H is indicative of peoples in the southern portion of the Indian sub-Continent and Bangladesh. Added now to these same peoples are Haplogroups L and T. Related to the peoples of South Asia are the Melanesian peoples of Southeast Asia and the Pacific who carry the additional Haplogroups M and S. All these peoples descend from Noah’s son Ham (with one exception) and their Haplogroups include:
[F], H, J, L, M, [P], S and T.
The exception being Canaan and his male descendants Haplogroups:
A, B and E.
Haplogroups C and D are associated primarily with Central Asians and East Asians, who descend from Noah’s eldest son Japheth. Added now to these are Haplogroups N, O and Q. Haplogroup Q being the defining marker Haplogroup for the Amerindian. Thus Japheth’s male descendants Haplogroups include:
C, D, [K], N, O and Q.
The intersection Haplogroups F and P are both found on the Indian sub-continent, while K is found in South East Asia.
Haplogroup G is the first ostensibly European Haplogroup followed by the later mutations from Haplogroup I and are indicative of Shem’s descendants, the second son of Noah. The Key addition to these are the relatively recent mutations of R1a and R1b. Thus the Haplogroups of the male descendants of Shem include:
G, I and R.
Therefore Haplogroups A, B, E1a, E1b1a, E1b1b and E2 are indicative of the offspring of Canaan; J1, the sons of Mizra and J2 of Phut; while Haplogroup H (and L) of the sons of Cush. Haplogroup C is located the most frequently amongst Madai in central Asia today and Haplogroup D in Tarshish, the second son of Javan. Haplogroup O is found in Gomer, Javan, Magog, Tubal and Meshech in East Asia and Q in Tiras, the native American Indian.
Haplogroup G is more difficult to isolate beyond Shem, whereas Haplogroup I is indicative amongst descendants of Shem’s third born son, Arphaxad. Haplogroup R, split into R1a is the marker in Arphaxad’s great grandson Joktan of Eastern Europe and in Asshur of Russia, and by degree, Lud in Iran; while R1b is the marker in Arphaxad’s great grandson Peleg of Western Europe, Aram in Latin Europe and Latino America, and by degree, Elam of Turkey.
While Haplogroups may indicate admixture through intermixing, integration and intermarriage and mutate accordingly, such as Haplogroup N of East Asian origin yet also found in high concentrations in the European Baltic nations and Finland; N remains an Oriental, Asian, Eastern line of descent from Japheth originally.
Likewise with major divisions in Haplogroups such as E1b1a and E1b1b, J1 and J2 and R1a and R1b, these are indicative of related peoples respectively from Canaan and in part Mizra for Haplogroup E; Mizra and Phut for Haplogroup J; and Joktan and Peleg for Haplogroup R.
As discussed regarding Haplogroups A to I, most of these Haplogroups, whether ancient or old have a lower frequency in the world with less mutations and include A, B, C, D, F and G. Haplogroup H though deemed old, is found in high concentrations, while contrastingly Haplogroup I is less concentrated with numerous sub-clades. It is Haplogroup E which stands out, as a widespread Haplogroup; one with high concentrations; and numerous mutations and sub-clades.
Considering the intermediate to younger Y-DNA Haplogroups which have a lower frequency in the world with less mutations they include K, M, P, S and T. They can be added to Haplogroups A, B, C, D, F and G. Haplogroups found in either relatively ‘high concentrations’ or with ‘numerous sub-clades’ include L, N and Q. They in turn can be added to Haplogroups H and I. This leaves Haplogroups J, O and R. Like Haplogroup E, they are indicative of being widespread; highly concentrated; with many mutations; and found in large population clusters. While E is old, Haplogroup J is intermediate and O and R are far more recent.
Haplogroup J mirrors E in two ways in that a. it splits into two, J1 and J2; and b. they broadly represent two different yet closely related peoples from Ham – Mizra and Phut. Haplogroup O finds community with E and J in that it has split into two main sub-Haplogroups O1 and O2, yet is a marker Haplogroup for nearly all of Japheth’s descendants. Finally, Haplogroup R has also split into two primary groups of R1a and R1b. Though perhaps more than Haplogroups E, J and O even, R1 represents 95% to 99% of Shem’s descendants.
Listing all the Y-DNA Haplogroups, results in a total of twenty:
A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, P, Q, R, S and T.
If we were to minus the Haplogroups one could term connecting Haplogroups, that is the ones which spawned each major set of mutations, then they would broadly include B(T), F, K and P. Thus lowering the total to sixteen. If we again subtract those Haplogroups which even though they may be dispersed over a wide geographic area, they remain smaller in regard to population numbers. These would include Haplogroups L, M, N, S and T. The total is now eleven. Of those remaining there are five which have split into two major sub-clades and they comprise Haplogroups E, I, J, O and R.
Thus, a configuration for major paternal lines as evidenced by Haplogroup groupings would be:
A, C, D, E1b1a, E1b1b, G, H, I1, I2, J1, J2, O1, O2, Q, R1a and R1b.
A total of sixteen, representing Noah’s grandsons and equating to the sixteen major ethnicities on the Earth. The expansion of the core Haplogroups into the myriad sub-clades today, is the scientific record of the story unfolding of a very small family grown incredibly large.
While this works for a conventional explanation of chapters nine and ten of Genesis, it is not accurate. As Canaan was not Ham’s son but his step son after an encounter between his wife Na’eltama’uk and Noah – refer Chapter XI Ham Aequator. Therefore we are seeking twenty-one grandsons from four sons of Noah. For Ham only had three sons and Canaan had six – refer Chapter XII Canaan & Africa.
If we were to categorise the paternal Haplogroups according to the number of Noah’s grandsons it would result if manipulated one way in five major Haplogroups representing Japheth’s sons. Though Japheth had seven sons in total, the grouping of Magog, Tubal and Meshech as one represents China (Chapter X China: Magog, Tubal & Meshech), with Tiras, Madai, Gomer and Javan – the Haplogroups being C, D, O1, O2 and Q, with Haplogroup C2 indicative of Central Asia; D1a in Japan; O in East Asia and South East Asia; and Q in the Americas.
If reshuffled another way, there are seven paternal Haplogroups which define East Asian (oriental) men, coincidently equaling the seven sons of Japheth. This configuration resulting in the Y-DNA Haplogroups C, D, K, N, O1, O2 and Q; though if one wished to add O1a and O1b, then possibly the interconnecting Haplogroup K could be omitted: C, D, N, O1a, O1b, O2 and Q. Perhaps this alignment is feasible and most accurate and conveniently squares with seven Haplogroups for seven grandsons.
Interestingly, the three sons of Ham are represented by the three major paternal Haplogroups consisting of H, J1 and J2, with H1a in South Asia, J1 in the Middle East and J2 in West Asia.
The major Y-DNA Haplogroups for Canaan include A, B and E. Haplogroup E can be split into E1b1a and E1b1b, making four. If manipulated further, Haplogroups E1a and E2 are bonafide enough to result in six Haplogroup lines, fascinatingly matching the six sons of Canaan – Sidon, Heth, Amor, Hiv, Arvad and Hamath. All Black African males fall into one of these six lineages, even though granted the majority possess E1b1a or E1b1b.
While Shem’s Haplogroups do not need any shuffling as his five sons correspond with the major Haplogroups G, I1, I2, R1a and R1b. Haplogroup G2a is indicative of the Caucasus; I1 in north western Europe; I2a1 in southeastern Europe; R1a in Eastern Europe; and R1b in Western Europe.
Thus the final configuration for paternal Y-DNA Haplogroups numbering twenty-one instead of sixteen would be:
A, B, C, D, E1a, E1b1a, E1b1b, E2, G, H, I1, I2, J1, J2, N, O1a, O1b, O2, Q, R1a and R1b.
Japheth: C, D, N, O1a, O1b, O2 and Q
Canaan: A, B, E1a, E1b1a, E1b1b and E2
Shem: G, I1, I2, R1a and R1b
Ham: H, J1 and J2
The Haplogroups in bold are dominant not just in geographic frequency but in geographic concentration as well. Three each for eldest son Japheth and third son Ham and two each for second born son Shem and the illegitimate youngest son of Noah, Canaan.
The field of genetics is fascinating and the facts being uncovered are of great interest. It would be enough for geneticists to stay with what is known, but as is scientist’s proclivity, it is the explanation of them where theoretical inaccuracies can be exposed. A case in point are scientists explanations of where a specific Haplogroup mutated or when. It includes considerable guesswork, yet often the terminology used, conveys the impression that the theories presented are factual.
Geneticists have elongated the chronological timeline for each mutation in support of the evolutionary theory. The reality, is that they should be concertinaed down to a considerably shorter time window as per an unconventional chronology – refer Appendix IV: An Unconventional Chronology.
Thus, a misdirection in studying Haplogroups is focusing on where and when a Haplogroup mutation originated. The key to understanding Haplogroups more clearly, is rather with whom did the mutation derive? Yet what record would provide this information?
To really grasp the meaning of the data, the answer amazingly, is in a book with a rather dusty cover, as not many people open it, or if they do, they perhaps do not know where to look or how to decipher it. As the constant reader will know, the identity of all the peoples and nations in the world are in the scriptures. In the book of Genesis in chapter ten there is what biblical scholars call the Table of Nations. A family tree if you will, of all the peoples in the world today. A 2021 genetic study highlighted that the many ethnicities on Earth can be placed into at least three broad categories, from which everyone descends – refer Chapter I Noah Antecessor Nulla.
On Earth today there are twenty-one major racial lines of descent; which have sprung from four original founding ancestor groups. These progenitors were Noah’s four sons, Japheth, Shem, Ham and Canaan, in the order with which they were born (Genesis 10:21), his twenty-one grandsons and their wives. The period for paternal and maternal Haplogroup evolution falling between the creation of Adam and Eve circa 27,000 BCE, the births of Noah’s three sons circa 12,000 BCE and the birth of Abraham circa 2000 BCE.
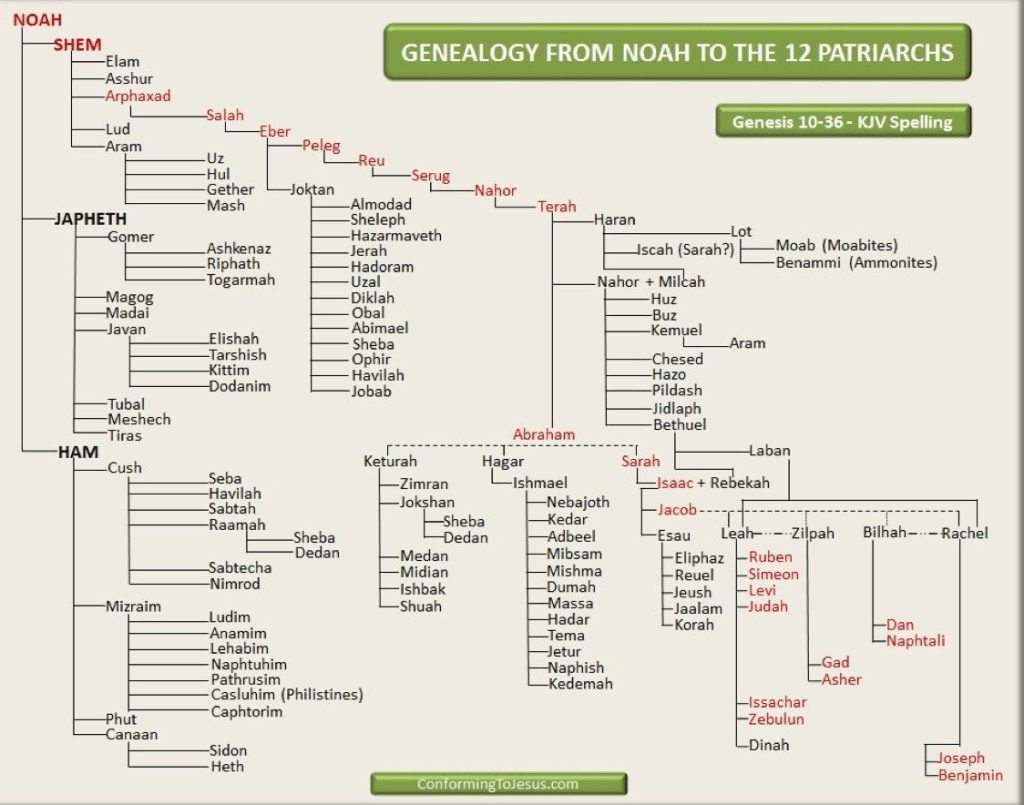
A universal misnomer is that Japheth is the progenitor of the European, Western, White peoples of the Earth; when in fact it is Noah’s son Shem. Broadly, the peoples descended from Noah’s four sons are the following.
A. Japheth: Central Asia, East Asia, Southeast Asia, Polynesia and the Amerindian – Genesis 10:2–5
B. Ham: North Africa, the Middle East, Arabia, South Asia and Melanesia – Genesis 10:6-7, 13-14
C. Canaan: Sub-Saharan Africa (and North Africa) – Genesis 10:15-19
D. Shem: Europe, West Asia, North America, Brazil, Latino-Hispano America, Australia, New Zealand and South Africa – Genesis 10:8-12, 21–31
The following are the major ethnic lines of descent, which have sprung from the four original founding ancestor groups:
A
1 Chinese
2 Japanese
3 Koreans
4 Continental South East Asians: for example, Vietnamese
5 Archipelago South East Asians: for example, Indonesians/Malays
6 Central Asians: for example, Kazakhstan/Turkic-Mongols
7 The Amerindian of North, Central and South America
B
1 Indians and related peoples: for example Sri Lanka
2 Pakistan
3 Arabs
C
1 Southern Africa
2 Central Africa
3 Western Africa
4 Eastern Africa
5 Horn of Africa
6 North Africa (Berbers)
D
1 Iranians/Persians
2 Turks
3 Latins: Portugal, Spain, Brazil and Latino-Hispano Americans: for example Mexico and Argentina
4 Russians
5a Eastern Europeans: Finns, Balts, Slavs, Balkans and Greeks
5b Western Europeans: British, Irish, Scandinavian, Benelux, Germans, French, Swiss and Italians

Ostensibly, twenty-two major ethnicities exist. Though for reasons the constant reader will be aware, the correct answer is twenty-one as Eastern Europeans and Western Europeans both descend from Shem’s third born son, Arphaxad.
It is worth mentioning that Italy contains a complex population demographic in that four major biblical lines of descent are included: from Abrahams’s brother Nahor and his wife Milcah (1) and Nahor with his concubine Reumah (2); descendants from one of Aram’s four sons, Uz (3); as well as peoples descended from one of Joktan’s thirteen sons, Uzal (4).
The principal Y-DNA Haplogroups for these twenty-one lines of descent; beginning with Japheth. The main group for Chinese men (Magog, Tubal and Meshech) is O at 82% and it is O2a in 56% of them which is dominant. For Japanese men (Tarshish) Haplogroup O represents 51% of the population, with O1b the highest at 30%. Though, the single biggest Haplogroup is D1a with 40%. In South Korean men (Togarmah) it is Haplogroup O with 79% and O2a the most dominant at 42%.

Another son of Gomer in South East Asia (Ashkenaz) are Vietnamese men who have Haplogroup O at 79%; with the dominant clade being O2a with 40%. Amongst other sons of Javan in South East Asia are Filipino men (Dodan) with 70% of Haplogroup O and O2a at 39%. In Malaysia (Elishah) it is Haplogroup O also at 70% and O1b edging O2a with 32% to 30% respectively. In Indonesia (Kittim), Haplogroup O stands at 69% and in Java O1b dominates with 42%; whereas in Sumatra, O2a is the biggest Haplogroup with 40%.
In the Central Asian nation of Kazakhstan (Madai) the dominant Haplogroup is C at 40%, while O is only 8%. Similarly in Mongolia, Haplogroup C is 51% and O, 16%. The North American Indian (Tiras) is defined by Q, with 77% of men carrying the Haplogroup and O virtually non-existent. In Javan’s descendants in Micronesia, Haplogroup K dominates in 65% of men and O at only 9%. Contrastingly in Polynesia, the Cook Islanders carry C as the highest Haplogroup, with 83% and O at only 5%.
Haplogroup O is by far the most dominant Haplogroup marker for Japheth’s male descendants. Of the three main sub-clades, it is O2a which is clearly the most frequent. Other Haplogroups in order, such as Q, C, D, K and N are far less numerous in frequency or in concentration compared with Haplogroup O. What is interesting about this is how relatively young or recent the O mutation is in comparison to other older Haplogroups. Haplogroups N and Q can be added as relatively recent too.
Haplogroup K is an intermediate intersection Haplogroup and though old, it is Haplogroups C and D which are legitimately called ancient. Those peoples who exhibit these Haplogroups more frequently, such as Kazakhs, Mongolians, Tibetans and the Japanese are reflecting an ancient lineage with incredibly less mutations. Reflecting endogamy and isolation perhaps. All the East Asians with Haplogroup O are showing a recent mutation stemming from C originally, yet having undergone a gigantic expansion relatively recently in humankind’s evolutionary history.
Ham would have originally carried similar genetic DNA with Japheth, yet the potential for different sets of mutations would have existed alongside his older brother. As each were the sons of Noah, who as ancestor zero was unique in history. Either, Noah’s genetic inheritance was manipulated in some fashion prior to his birth or, he simply carried what was common to the line of Seth in the first place – Chapter I Noah Antecessor Nulla. In any case the original Y-DNA Haplogroup A with its close descendant Haplogroup B, have remained lesser markers for Noah’s youngest son Canaan. Even so, in Namibia Haplogroup A found in 64% of males, compares with Haplogroup B found in 20% of the Zulu men in South Africa.
Haplogroup E1b1b is carried by 63% of Ethiopian men. The dominant Haplogroup in sub-Saharan Africans is Haplogroup E, with E1b1a having the highest frequency. For men in Ghana it is as high as 92%. The link between Canaan’s descendants throughout Africa is revealed in the shared Haplogroup E1b1b. It is a defining marker Haplogroup for the Berbers in the Arab world. In Morocco, it is found in 83% of the male population.
As one heads east, E1b1b decreases and Haplogroups J1 and J2 from Ham increase. Haplogroup J1 dominates the Arabian Peninsula and Middle Eastern Arabs, with Yemen men carrying 73% Haplogroup J1. Haplogroup J2 is the link between the Arab world descended from Mizra and the peoples of Pakistan from Phut. Haplogroup J2 has spilled over into the Levant and the Middle East, where 26% of Lebanese men exhibit J2 for instance.
In Pakistan, Haplogroup J2 is logically the true ancestral paternal Haplogroup and found in 20% of Pakistani men; rather than the Eurasian R1a Haplogroup from admixture. The descendants of Phut’s brother Cush, carry J2 from intermixing, though it is Haplogroup H and to a lesser degree L which are the true ancestral lineages for Cush’s sons. The highest concentration of Haplogroup H is found in Bangladeshi men with 36%; then India with 23%; Sri Lanka with 15%; and Pakistan with 6%. The highest percentage of Haplogroup L is found in Sri Lankan men at 19%; then India at 18%; Pakistan with 12%; and Bangladesh with 5%.
Haplogroups M, T and S as discussed, are found in small quantities or isolated geographic regions. Thus the core paternal Haplogroups for Ham’s descendants include the old to intermediary Haplogroups comprising H, J1, J2 and L. Melanesians also exhibit old to intermediary mutations in Haplogroups M and S (aka K2b1).
The obvious question, is why would a younger brother carry older Haplogroup mutations? One answer is that it is through Noah’s illegitimate youngest son Canaan, that the pre-flood genetics as typified by the paternal Haplogroup A0 were retained by Canaan’s descendants, even though once the ancestral Haplogroup for all twenty-one grandsons. This genetic inheritance has remained strong in Canaan’s line. Perhaps and likely, Canaan physically took after his mother Na’eltama’uk more than her other sons, Cush, Phut and Mizra – refer Chapter XI Ham Aequator.
Whatever the reason, it is undoubtedly Canaan who has retained the original A Haplogroup and its sub-clades, which began with Adam, passed to his third son Seth, to righteous Enoch and finally to Noah. Black Hebrew Israelites claim African Americans descend from ancient Israelites. This is inaccurate, having incorrectly appropriated a white line of Shem as their own; while at the same time disdaining any link with the line of Canaan. The irony, is that sub-Saharan Africans not only descend from Canaan, but as Canaanites, they embody the original line of lineal descent from Adam to Noah. To be a physical descendant of Abraham (b. 1977, d. 1802 BCE), one would be required to possess a ‘recent’ Haplogroup mutation some 4,000 years old. Whereas Haplogroup R1b-U106 fits the bill, Haplogroups A, B and E certainly do not.
While Canaan’s male descendants retain the ancient groups A, B and E, and Japheth’s retain the ancient groups C and D, the oldest significant Haplogroup for Shem’s descendants is G. Not quite ancient but the oldest of the groups before the intermediary Haplogroups, comprising I, J, K, and LT. Haplogroup G is found predominantly in the Caucasus region and does not equate specifically to the six sub-groups of Shem’s descendants as outlined earlier in Group C.
The same applies to Haplogroup I and its major mutations, I1 in the Nordic countries and I2 in the Balkans. It is only when we arrive at the recent Haplogroup mutation labelled R1 that we can appoint specifically the European peoples. The other recent Haplogroups comprise N, O and Q. Of Shem’s five sons, the split between R1a and R1b is it seems, exactly half way with two and a half sons each.
Iran has a very complex ethnic demographic – refer Chapter XVII Lud & Iran. This was foretold in the scriptures, for not just Lud but also Phut and Cush, where it says in Ezekiel 30:5 KJV: “[India] and [Pakistan], and [Iran], and all the mingled people…” The original Hebrew words are: Cush, Phut and Lud. In other words, India, Pakistan and Iran. As we have discovered in India and Pakistan, both have an array of paternal Haplogroups and admixture with peoples of Arabic and Eastern European stock. Iran is different in that the Persians are not descended from Ham but Shem. Though Iranians are similar in that they have a widespread number of Haplogroups, with none being overly dominant. Revealing heavy admixture as a result of their geography.
There are a percentage of Iranian men with Japheth’s Haplogroups Q (5.5%) and N (1%). Haplogroups indicating mixture with Mizra’s sons Ludim, Lubim and others are revealed in part with J2 (23%) and principality with J1 (8.5%) and E (6.5%). Similar descent from Ham is included in Haplogroups L (6.5%) and T (3%). Haplogroups indicating a common descent with the rest of Europe include the ancestral lines of G (10%) and I (0.5%). Haplogroup R1b is held by 9.5% of men and R1a in 15.5%. Comparing Iran with other sons of Shem it appears that R1a is the marker Haplogroup for Iranian men.
The Turks who descend from Elam, also possess a complicated demographic with an equal array of diverse paternal Haplogroups – refer Chapter XVIII Elam & Turkey. And again indicative of heavy admixture influenced by their geographic location. Haplogroups Q and N account for 6% of men, while J1 (9%), J2 (24%), E1b1b and T account for 46.5%. The lineages of Shem, G and I add up to 16.5%. Turkish men who carry R1a account for 7.5% of the population and those who possess R1b, 16%. Turkey is a mirror opposite to Iran regarding R1a and R1b. Thus the defining Haplogroup for Turkish men appear to be R1b.
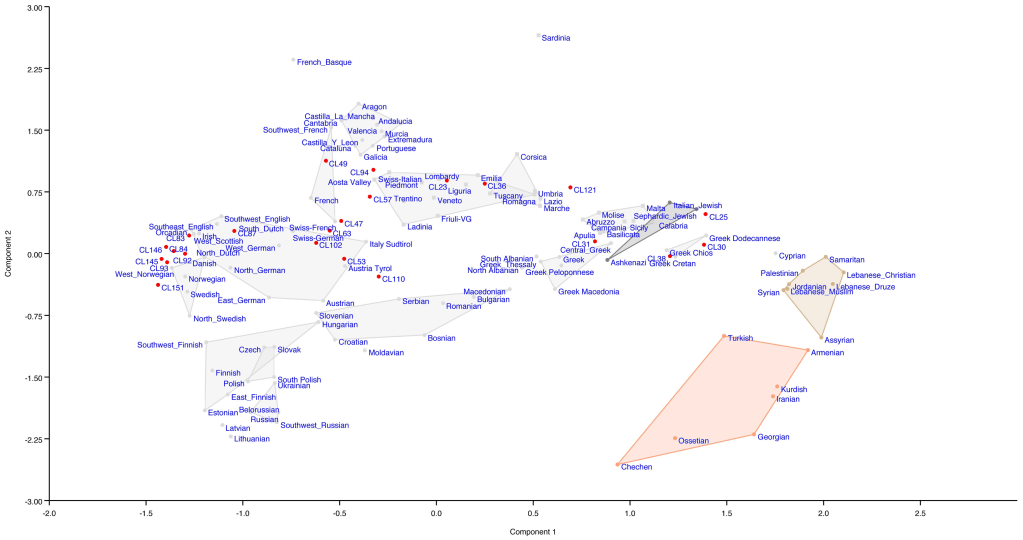
The Latin peoples of southern and western Europe and their descendants in the Americas are all dominant in R1b and descend from Aram – Chapter XV The Philistines: Latino-Hispano America; and Chapter XXIII Aram & Tyre: Spain, Portugal & Brazil. In Mexico, the R1b percentage is 50%; Brazil, 54%, Portugal, 56%; and Spain, 69%.
The Eastern Europeans descend from Arphaxad’s great grandson Joktan and their primary Haplogroup is R1a as it is for the Russians from Asshur – Chapter XX Will the Real Assyria Stand Up: Asshur & Russia; and Chapter XXIV Arphaxad & Joktan: Balts, Slavs & the Balkans. Russian men have 46% R1a and in Poland it is 58%, the highest in Europe. Which leaves the Western Europeans, the family of Abraham and his brothers who all descend from Arphaxad’s great grandson Peleg and where the defining Y-DNA Haplogroup is R1b. The highest incidence of R1b in males is in Ireland with 81%, though some figures give Welsh men, 92%.
Similarly with Japheth’s descendants who have less defining Haplogroups (O) than Ham’s descendants (H, J1, J2); Shem’s descendants also are almost entirely defined by relatively recent, younger Haplogroups. In this case, R1a and R1b. If we were to ascribe a different alphabetic code for paternal Haplogroups as pertinent to N for Noah, J for Japheth, H for Ham, C for Canaan and S for Shem, then the mutational sequence would look something like this.
A – N
B – C
C – J
D – J1
E – C1a, C1b
G – S
H – H
I – S1a, S1b
J -H1a, H1b
L – H2a
T – H2b
M – H3a
S – H3b
N – J2
O – J3a, J3b
Q – J4
R – S2a, S2b
This sequence highlights the fact that the quantity of Haplogroups are staggered with the most numerous for firstly Japheth, closely followed by Ham; thirdly for Shem and fourthly Canaan. Linked with this, are the number of increased mutations associated with Ham and Canaan compared with either Japheth or Shem.
Canaan’s descendants paternal Haplogroups have been impacted by primarily inheriting the genetic similarity of antediluvian humankind; while Japheth’s descendants have likewise been affected by a genetic inheritance, this time influenced by Neanderthal and Denisovan DNA – refer articles: Homo neanderthalensis I, II, III & IV; and Chapter XXII Alpha & Omega.
These are both ancient lines and so it shouldn’t be a surprise that Shem’s DNA received from Noah who happened to look different from everyone else, is recessive and mutated after the other two pre-existing lineages which crossed over into the post-flood world – refer Chapter I Noah Antecessor Nulla; and Chapter XVI Shem Occidentalis.
Considering mtDNA maternal Haplogroups and Y-DNA paternal Haplogroups, sub-Saharan Africans for example have inherited an older sequence of genetic material than Indians and Arabs; Indians and SArabs an older set than East Asians and East Asians an older set than Europeans. Black people also possess the widest variation in their genome than any other people on the planet. In fact, there is more variation amongst the peoples of Africa than the rest of the world combined.
Europeans are the bridge between Black Africans and East Asians. Between Europeans and the sub-Saharan Africans are the remaining peoples descended from Ham, the Arabs and Indians.

A principal component analysis (or PCA) graph, confirms the association of these four ancestral groups. Millenniums of mutations have resulted in the East Asians being the most distantly separated from Africans.
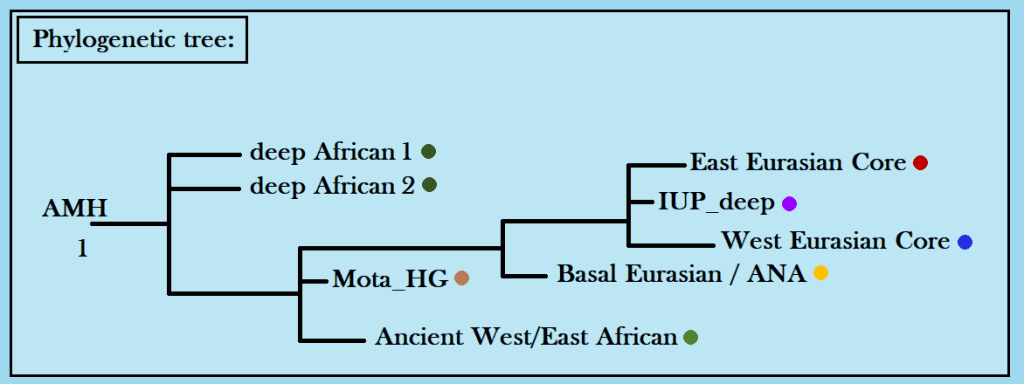
The fact that East Asians possess the highest percentages of Neanderthal DNA and Black people possess virtually none is symptomatic of their contrasting genetic relationship.
This study poses more questions than answers. Why have paternal Haplogroup mutations followed the patterns they have for Noah’s four sons and twenty-one grandsons? Why for instance, does the fifth grandson son of Japheth, the second son of Javan, Tarshish and the Japanese have a high percentage of Haplogroup D1a compared to all other descendants from Japheth? Why does Tiras the Amerindian, the youngest son of Japheth have a markedly different Haplogroup (Q) from his brothers who all share principally O?
Why are Canaan’s descendants the Black Africans so obviously different in physiognomy and Haplogroups (MtDNA L0-L6 and Y-DNA, A, B and E) from their brothers, the Arabs, Pakistanis and Indians?
This question was certainly a puzzle until it became clear that Canaan is their half brother, sharing the same mother with Mizra, Phut and Cush but not the same father.
Even so, the pressing question remains in how and why Noah’s youngest son has retained the oldest paternal Y-DNA Haplogroups. Not to mention his descendants possessing the oldest mtDNA Haplogroups as well.
This article is not a definitive study by any stretch and is very much a work in progress. Particularly as this writer’s knowledge and understanding of Haplogroup lineages grows with additional research.
God began by making one person, and from him came all the different people who live everywhere in the world. God decided exactly when and where they must live.
Acts 17:26
New Century Version
© Orion Gold 2023 – All rights reserved. Permission to copy, use or distribute, if acknowledgement of the original authorship is attributed to Orion Gold
Post Scriptum
Originally this addition was to correct an error, now rectified in the main body of the article. Though it is worth retaining as per below for the important point it presents.
An error which requires correction is that of Y-DNA Haplogroup E.
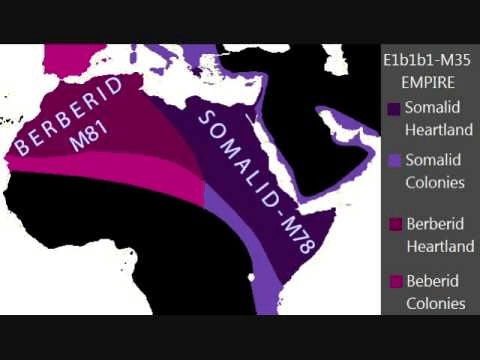
As the map above shows, E1b1b is indicative of North African and East African men and not of Arab related males in the same regions who carry J1 or J2.

In other words, Berber and Ethiopian men who carry E1b1b are more closely related in paternal lineage with men of sub-Saharan Africa who possess E1b1a, E1a or E2.

Addendum I
The information following, lends support against the Out of Africa hypothesis as discussed here and in other articles. It also highlights the difference between Black Africans and White Europeans, in that the latter may not have descended from the former as paternal Haplogroups have indicated through their alphabetisation sequence. Lastly, it corroborates our ancestor being Cro-Magnon man and tellingly, approximately 30,000 years ago – refer Chapter I Noah Antecessor Nulla; Chapter XXII Alpha & Omega; and Appendix IV: An Unconventional Chronology.
“Out of Africa” Theory Officially Debunked, Robert Sepehr, May 3, 2014 – emphasis mine:
‘Scientific evidence refuting the theory of modern humanity’s African genesis is common knowledge among those familiar with the most recent scientific papers on the human Genome, Mitochondrial DNA and Y-chromosomes. Regrettably, within mainstream press and academia circles, there seems to be a conspicuous – and dare we say it – deliberate vacuum when it comes to reporting news of these recent studies and their obvious implications.
Australian historian Greg Jefferys explains that, “The whole ‘Out of Africa’ myth has its roots in the mainstream academic campaign in the 1990′s to remove the concept of Race. When I did my degree they all spent a lot of time on the ‘Out of Africa’ thing but it’s been completely disproved by genetics. Mainstream still hold on to it.”
It did begin the early 90’s. And the academics most responsible for cementing both the Out-of Africa theory and the complementary common ancestral African mother – given the name of “Eve” – in the public arena and nearly every curriculum, were Professors Alan C. Wilson and Rebecca L. Cann. In their defense, the authors of this paper were fully aware that genealogy is not in any way linked to geography, and that their placement of Eve in Africa was an assumption, never an assertion.
A… paper on Y-chromosomes published in 2012, (Re-Examining the “Out of Africa” Theory and the Origin of Europeoids (Caucasians) in the Light of DNA Genealogy written by Anatole A. Klyosov and Igor L. Rozhanski) only confirms the denial of any African ancestry in non-Africans, and strongly supports the existence of a “common ancestor” who “would not necessarily be in Africa. In fact, it was never proven that he lived in Africa.”
Central to results of this extensive examination of haplogroups (7,556) was the absence of any African genes. So lacking was the sampling of African genetic involvement, the researchers stated in their introduction that, “the finding that the Europeoid haplogroups did not descend from “African” haplogroups A or B is supported by the fact that bearers of the Europeoid, as well as all non-African groups do not carry either SNI’s M91, P97, M31, P82, M23, M114, P262.”

‘With the haplogroups not present in any African genes and an absence of dozens of African genetic markers, it is very difficult nigh on impossible to sustain any link to Africa. The researchers are adamant that their extensive study “offers evidence to re-examine the validity of the Out-of-Africa concept”. They see no genetic proof substantiating an African precedence in the Homo sapien tree, and maintain that “a more plausible interpretation might have been that both current Africans and non-Africans descended separately from a more ancient common ancestor, thus forming a proverbial fork”.
We regard the claim of “a more plausible explanation” as a gross understatement, since there is absolutely nothing plausibly African turning up in any test tubes. In fact, the researchers made note of their repeated absence stating “not one non-African participant out of more than 400 individuals in the Project tested positive to any of thirteen ‘African’ sub-clades of haplogroup A”. The only remaining uncertainty relates to the identity of this “more ancient common ancestor”. All that can be stated with confidence is that humanity’s ancestor did not reside in Africa.
Unfounded accusations of racism have become common as the prevailing Afrocentric hypothesis is constantly being challenged by the growing mountain of conflicting scientific evidence, especially in the evolving field of genetics. It is now scientifically irrefutable fact that the “human species” has been found to contain a substantial quantity of DNA (at least 20%) from other hominid populations not classified as Homo sapien; such as Neanderthal, Denisovan, African archaic, Homo erectus, and now possibly even “Hobbit” (Homo floresiensis).
If not given drugs to prevent infant death, the pregnant body of a rhesus negative mother will attack, try to reject, and even kill her own offspring if it is by a rhesus positive man. The Domestic dog (Canis lupus familiaris) is a sub-species of the gray wolf (Canis lupus), and they produce hybrids. There are numerous other examples of where two separate species (for example with different numbers of chromosomes) can also produce viable offspring, yet are considered separate species.
That said, humanity has been shown to be, genetically speaking, a hybrid species that did not all share the same hunter-gatherer ancestry in Africa. Recent sequencing of ancient genomes suggests that interbreeding went on between the members of several ancient human-like groups more than 30,000 years ago, including an as-yet unknown human ancestor. “there were many hominid populations,” says Mark Thomas, evolutionary geneticist at University College London.’

Addendum II
The idea that the sequential, chronological age of paternal Y-DNA Haplogroups may correlate with the births of Noah’s grandsons is an intriguing one.
Comparing the sons of Ham, results in a perfect match with the order of birth and the age of their prime Y-DNA Haplogroup:
Cush – H
Mizra – J1
Phut – J2
Regarding Canaan’s sons there doesn’t seem to be any correlation apart from perhaps with Sidon and Hiv. Though as E1b1a, E1b1b and A are the predominant African Haplogroups, it would need to be investigated further and thus an alignment may yet be found.
Sidon – A
Heth – B
Amor – E1a
Hiv – E1b1a
Arvad – E1b1b
Hamath – E2
Shem’s sons all carry R1a or R1b as their prime Haplogroup. Their order of birth in Genesis is as follows below, but if Elam and Lud were swapped and Peleg and Joktan as well, it would be 100% accurate. One wonders if the biblical order for Shem’s sons is correct?
Elam – R1b
Asshur – R1a
Arphaxad
Peleg – R1b
Joktan – R1a
Lud -R1a
Aram – R1b
The new order would be: Lud, Asshur, Joktan, Peleg, Elam and Aram.
The sons of Japheth are perhaps the most complex, but if we stay with the prime Y-DNA Haplogroup (regardless of percentages for other Haplogroups from integration) then the results are the following.
Gomer
Ashkenaz – O1b
Riphath – O1b
Togarmah – O1b
Magog – O2
Madai – C
Javan
Elishah – O1b
Tarshish – O1b
Kittim – O1b
Dodan – O1a
Tubal – O1a
Meshech – O1b
Tiras – Q
Tiras as the youngest son correctly possesses the most recent Haplogroup mutation for Japheth: Haplogroup Q.
Magog (O2), Tubal (O1a) and Meshech (O1b) who are all males in China each carry a different mutation for Haplogroup O. Apart from the order – though scientists admit the O1 and O2 mutations could have been simultaneous – actually confirms the tripartite component in China’s population as discussed in Chapter X China: Magog, Tubal & Meshech.
The sons of Gomer all carry O1b, as do the sons of Javan. With the exception of the Philippines (Rodan) which exhibits a low percentage of O1b and is a bit of an anomaly anyway.
Madai caught in the middle (as his name means) with Haplogroup C runs against the pattern, though with the exception of Madai (and Rodan), the descendants of Japheth follow an almost perfect pattern like his brother Ham.